Analysis of antimicrobial resistance and class 1 integrons among strains from upper respiratory tract of healthy adults
Introduction
Upper respiratory tract infections (URTIs) and community-acquired pneumoniae (CAP) are highly prevalent diseases of respiratory system associated with significant morbidity and socioeconomic cost. Because the nasopharynx lies between the nose, sinuses, ears, larynx, and the lower respiratory tract, resident pathogens of the nasopharynx can be the source for both upper and lower respiratory tract infections (1,2). The nasopharynx is also a major source of secretions containing bacteria that can easily spread between individuals and these may subsequently become pathogenic in the new host. In this study, the issue of nasopharyngeal carriage is investigated, because it plays an important role in both the development of disease and the spread of pathogens. In most cases these nasopharyngeal flora are carried without causing clinical symptoms. However, when homeostatic conditions of the host are altered, microorganisms may invade adjacent sites causing disease. Considering studies isolates to antimicrobial susceptibility in China have largely focused on isolates recovered from clinical specimens in the context of clinical disease, in this study, the antimicrobial susceptibility of the major opportunistic pathogens from healthy adults was also performed.
Integrons are ancient structure that contains determinants of a site-specific recombination system to capture genes encoding antimicrobial resistance (3). They can locate within transposons or conjugative plasmids and contribute to the traffic leading to the acquisition of new genes in bacteria (4-8). The class 1 integron is prevalent on plasmids. It has been found in both gram-positive and especially in gram-negative bacteria (8-11). The study of integrons most focus on clinical strains, few on nasopharyngeal flora from healthy adult. In this study, we investigated the distribution and characterization of the integrons among opportunistic pathogens from healthy adults.
Materials and methods
Subjects
A total of 1,019 nasopharyngeal samples from healthy adults were collected during the period July 2009-July 2010. 1,019 volunteers (n=1,019; 661 male; 358 female; mean 50±17 years), randomly selected from registry of general practitioners in Nanjing without symptoms or signs of clinical illness were enrolled in the study over a six-month time period and all of them gave their consent for the participation in this study. The healthy volunteers included had not been following any history of respiratory tract infection, chronic basic lung disease, had not received antibiotic treatment for at least six months prior to inclusion and did not have any relation with the hospital environment.
Nasopharyngeal samples
Samples were collected from both anterior nasopharynxes rotating a sterile polyester fiber-tipped swab moistened with sterile saline. Swabs were placed in 3 mL of Luria-Bertani broth and transported to the Department of Clinical Microbiology of the first Affiliated Hospital of Nanjing Medical University. All nasopharyngeal samples were inoculated on blood agar plates and chocolate agar plates (bioMerieux, France) and identified by the analytical profile index procedure (API-20NE system; bioMerieux, France).The reference strains used in this study were as follows: Staphylococcus aureus ATCC 25923, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853.
Antimicrobial sensitivity testing
Antimicrobial sensitivity testing was performed by the disc diffusion (Kirby-Bauer) method for Methicillin-resistant Coagulase Negative Staphylococci (MRCoNS), Methicillin-resistant Staphylococcus aureus (MRSA), K. pneumoniae, Haemophilus according to Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2010). The isolates were interpreted as susceptible or resistant according to the inhibition zone diameter using CLSI recommendations. Discs were obtained from Oxoid. The antimicrobials used for the susceptibility testing were as follows (μg per disc): fusidine (5 μg), penicillin (10 μg), oxacillin (1 μg), cefoxitin (30 μg), gentamicin (10 μg), rifampin (5 μg) ciprofloxacin (5 μg), levofloxacin (5 μg), moxifloxacin (5 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), azithromycin (15 μg), erythromycin (15 μg), clindamycin (2 μg), chloramphenicol (30 μg), quinupristin/dalfopristin (15 μg), tetracycline (30 μg), amoxicillin/clavulanic acid (30 μg), cefoperazone/sulbacta (30 μg), piperacillin/tazobactam (100/10 μg), cefazolin (30 μg), cefuroxime (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), cefepime (30 μg), aztreonam (30 μg), imipenem (10 μg), meropenem (10 μg), amikacin (30 μg), ampicillin (10 μg), ampicillin/sulbacta (10/10 μg), Antimicrobial susceptibility test of MRCoNS, MRSA and Streptococcus pneumoniae was performed using the E-test (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar (Oxoid, UK). The values of the minimal inhibitory concentration (MIC) of MRSNS/MRSA isolates to vancomycin and the values of the MIC of Streptococcus pneumoniae isolates to erythromycin, clindamycin, penicillin, vancomycin, levofloxacin, ampicillin, and cefotaxime were determined according to the manufacturer’s instructions and to CLSI guidelines.
Detection of integrons
To study the distribution and characterization of the integron among opportunistic pathogens from healthy adults, all isolates were screened for class 1, 2 and 3 integrons by PCR using degenerate primers hep35 (5′ TGCGGGTYAARGATBTKGATTT 3′) and hep36 (5′ CARCACATGCGTRTARAT 3′) and Hinf I restriction analysis of the integrase gene product (12). Cassette regions of class 1integron were amplified using primers 5′CS and 3′CS as described previously (13). Cassette PCR products were sequenced. The resulting DNA sequences were analyzed by the BLAST program, available at the NCBI homepage (
Statistical analysis
We analyze the difference of isolated rates between different age groups. The statistical analyses were performed using SPSS 17.0 software programs, employing the chi-aquare test. P values less than 0.05 were considered statistically significant.
Results
Bacteria group distribution
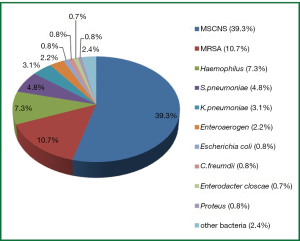
Out of the 1,019 samples, 743 (72.9%) opportunistic pathogens were isolated. The top five strains were as follows: Coagulase-negative staphylococcus (CoNS, n=404), S. aureus (n=109), H. influenzae (n=74), S. pneumonia (n=49) and K. pneumonia (n=32), other opportunistic pathogens such as Escherichia coli, Enteroaerogen were also detected (Table 1, Figure 1).
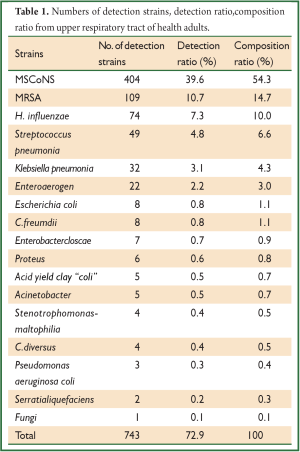
Full table
Comparing different age groups, we found that the isolated rates of S. aureus and H. influenzae were decreased with aging (Table 2). The isolated rates of S. aureus and H. influenzae age group of 41-60 (Staphylococcus aureus, 10.0%; H. influenzae 5.9%) and age group of 61-90 (S. aureus, 7.3%; H. influenzae 4.2%) compared to age group of 19-40 (S. aureus, 14.8%; H. influenza 11.0%) were different (P<0.05).
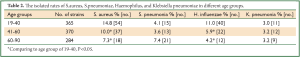
Full table
Antimicrobial susceptibility profiles
Antimicrobial susceptibility of MRCoNS and MRSA isolates
A total of 162 MRCoNS isolates and 19 (MRSA) isolates of healthy adults origin obtained during the period 2009-2010 in Nanjing were tested for antibiotic susceptibility (Table 3). Of these, 100.0% was found to be fusidine and vancomycin susceptive, 96.9% was found to be rifampin susceptive. The susceptive rate for MRCoNS and MRSA of other antibiotic was lower. Of these, 100.0% was found to be penicillin resistant. Overall, the susceptive rate of MRSA was lower than the susceptive rate of MRCoNS.

Full table
Antimicrobial susceptibility of S. pneumoniae isolates
A total of 49 S. pneumoniae isolates were tested for antibiotic susceptibility by testing the values of MIC. Most of these isolates were high resistant rate to erythromycin (71.2%), clindamycin (76.9%), and penicillin (74.4%). None isolate was found to be vancomycin and amoxicillin resistant (Table 4).
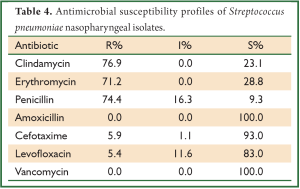
Full table
Antimicrobial susceptibility of K. pneumoniae isolates
Out of 32 K. pneumoniae isolates, 8 (25.0%) isolates were ESBLs-producing K. pneumoniae isolates. Overall, antimicrobial resistant rate of K. pneumoniae isolates was at a low level (Table 5).
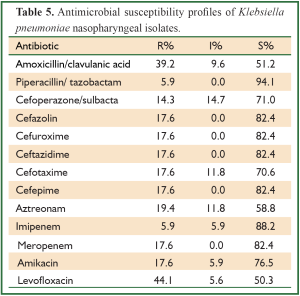
Full table
Antimicrobial susceptibility of H. influenzae isolates
A total of 74 of H. influenzae isolates were tested for antibiotic susceptibility (Table 6). None isolates was found to be ampicillin/sulbacta, piperacillin/tazobactam, cefazolin, cefuroxime resistant, 44.4% of isolates were trimethoprim-sulfamethoxazole resistant. Table 6 shows antimicrobial susceptibility of H. influenzae isolates.
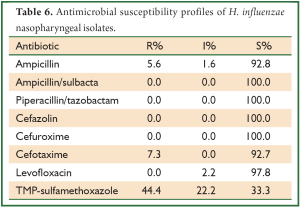
Full table
Prevalence of class 1 integrons
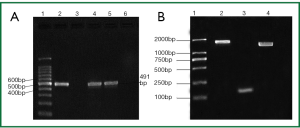
We found a low rate of class 1 integrons (0.4%) among 743 opportunistic pathogens of healthy adults origin obtained during the period 2009-2010 in Nanjing. Out of the 32 K. pneumoniae isolates and 6 Proteus isolates, 2 (6.3%) K. pneumoniae isolates and 1 (16.7%) Proteus isolates carried class 1 integron genes (Figure 2A). One of the gene cassette positive K. pneumoniae is an ESBL-producer. Class 1 integrons were not detected in any gram-positive strains and any other gram-negative strains. No class 2 and 3 integrons were detected.
DNA sequence analysis
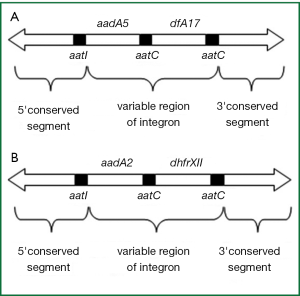
Among intI1-positivestrains, an amplicon of 1,584 bp was yieded in K. pneumoniae isolates, an amplicon of 1,974 bp was yieded in Proteus isolates (Figure 2B). Sequencing analysis revealed that amplicon 1,584 bp (Nanjing v3) harbored gene cassette aadA5 and dfrA17 (Figure 3A), amplicon 1,974 bp (Nanjing v1) harbored gene cassette aadA2 and dhfrXII (Figure 3B), There was no class 1 integron found in gram-positive strains and any other gram-negative strains (Table 1).
Discussion
Bacterial resistance to antibiotics represents one of the most significant global health challenges of this century. Infections with multiple antibiotic-resistant bacteria have been increasing at an alarming rate. Many infections caused by isolates occur in persons with prior nasopharynx carriage such as S. aureus infections (14), and this carriage is an important risk factor for healthcare-associated infections. Antibiotic and vaccines modify the flora by removing organisms that are part of the commensal flora. Therefore, we investigated the nasopharynx carriage status of opportunistic pathogens and test the antimicrobial resistance of major opportunistic pathogens from healthy adults for guiding of empirical therapy and for focusing interventional control of antimicrobial resistance in different geographic areas.
In this study, the major isolates was Gram-positive bacteria, the five most common organism identified isolates were CoNS, Staphylococcus aureus, H. influenzae, S. pneumoniae and K. pneumoniae. However, it was different to the distribution of clinical flora. In clinical isolates (15), Gram-negative bacteria accounted for 70.0%, the major isolates were E. coli, K. pneumoniae, P. aeruginosa, S. aureus and A. baumannii. It presents those antibiotics empirical treatment should be according to different sources of patients. This study also showed that nasal carriage rate of H. influenza and S. aureus varied with age (Table 2). H. influenzae typically asymptomatically colonize the nasopharynx of young children. To our knowledge, nasal carriage rate of H. influenzae and S. aureus varied with age may be relevant to immunity.
ESBLs have been detected in a wide variety of Gram-negative bacteria. K. pneumoniae is still an important ESBLs producer, not only in the nosocomial setting but also in the community (16). In this study, 8 (25.0%) ESBLs-producing K. pneumoniae were found from 32 K. pneumoniae isolated from the nasal carriage in healthy adults in Nanjing. We should be alert to the result that ESBLs-producing K. pneumoniae were found from healthy adults although the detection rate was at a low level.
In recent studies, 59.4% CoNS is more prevalent than S. aureus in clinical gram-positive Staphylococcus isolates in China (17-22). This study revealed that 69.0% of nasopharynx carriage status of opportunistic pathogens isolates was Staphylococcus isolates, including 54.3% of CoNS and 14.7% of S. aureus. Our surveillance showed that resistant rate of MRSA was higher than resistant rate of MRCoNS.
Nasopharyngeal carriage of S. pneumoniae may occur in up to 60.0% of healthy preschool children and up to 30.0% of older children and adults (23). In this study, it was up to 49 (4.8%) of S. pneumoniae isolates from healthy adults nasopharyngeal samples. Because the distribution of S. pneumoniae varies with age, geographic region and time (24), it was different nasopharynx carriage status of Streptococcus pneumoniae. Beta-lactams antibiotics and macrolides antibiotics were used for empirical therapy of CAP because the main strain of CAP was S. pneumoniae. However, resistant to penicillin and other antibiotics has increased dramatically worldwide over the past decades due to inappropriate antibiotic usage (25). Antibiotic resistance was commonly observed with beta-lactams and macrolides in Asia. The Asian Network for Surveillance of Resistant Pathogens (ANSORP) documented very high prevalence rates (>60.0%) of Streptococcus pneumoniae in Taiwan, Korea, Japan and Vietnam during 1996-1997 (26). In this study a trend towards increasing resistance to penicillin (74.4%), erythromycin (71.2%) and clidamycin (76.9%) was also seen.
H. influenzae is the second main pathogens in CAP. Resistance to ampicillin was first reported in the 1970s, and during the subsequent decades it has steadily increased. The resistance to ampicillin and other beta-lactam antibiotics is usually due to the production of a plasmid-encoded beta-lactamase, TEM-1 or ROB-1 (27). In Romania, 26.0% of H. influenzae stains isolated from patients with community acquired respiratory tract infections were resistant to amoxicillin (28). In France, 17.0% and 0.5% of H. influenzae stains isolated from the nasal carriage in children 3 months to 3 years of age were resistant to amoxicillin/clavulanic acid and cefotaxime (29). In this study, 5.6% and 7.3% H. influenzae stains isolated from the nasal carriage in healthy adults were resistant toampicillin and cefotaxime. In total, the resistant rate of H. influenzae was at a low level.
Of 32 K. pneumoniae isolate, 44.1% and 39.2% were resistant to Levofloxacin and amoxicillin/clavulanic acid, but 94.1% and 88.2% were susceptive to piperacillin/tazobactam and imipenem. It showed that we can choose piperacillin/tazobactam and imipenem as the first choice for empirical treating community-acquired K. pneumoniae infection.
Integrons are natural highly efficient recombination and expression systems able to capture genes as part of genetic elements known as gene cassettes (30). In recent studies, class 1 integrons were investigated from animals, water, human stools (31-34), however, to the best of our knowledge, this is the first report focusing on prevalence of class 1 integrons of strains from upper respiratory of healthy adults. In this study we investigated the occurrence, distribution, and cassette content of class 1 integrons among all 743 opportunistic pathogens of healthy adults origin obtained during the period 2009-2010 in Nanjing, China. No integron was found in gram-positive bacterium. Of Gram-negative bacterium, out of the 32 K. pneumoniae isolates and 6 Proteus isolates, 2 (6.3%) K. pneumoniae isolates and 1 (16.7%) Proteus isolates carried class 1 integron genes. In clinical Gram-negative isolates, Martinez,s study showed that integrons positive rate was 43.0% (5). Resistant gene cassette aadA5-dfrA17 (1,584 bp) was found in class 1 integron positive K. pneumoniae isolate, and aadA5-dfrA17 encoded aminoglycoside resistance gene and trimethoprim resistance gene. Resistant gene cassette aadA2-dhfrXII in class 1 integron positive Proteus isolate and aadA2-dhfrXII encoded aminoglycoside resistance gene and sulfonamides resistance gene. Although it was lower class 1 integron positive rate of strains from healthy adults than isolates associated with the clinical setting, class 1 integron harboring multidrug-resistance genes has been identified among strains isolated from upper respiratory of healthy adults. The results stressed the need for continued surveillance of bacteria from the asymptomatic carriers.
In conclusion, with a new generation of antimicrobial appearance and extensive use, respiratory infections become one of the most common infections. Abuse of antimicrobial agents causes respiratory tract infection pathogens change and increase. Therefore, it was essential to monitor resistance of common pathogen from community-acquired infections and dynamically analyze pathogen distribution and resistance of the respiratory infection. Moreover, in this study we found class I integron harboring multidrug-resistance genes among strains isolated from upper respiratory of healthy adults. The results stressed the need for continued surveillance of bacteria from the asymptomatic carriers.
Acknowledgements
We would like to thank Yang Ye and Chen Yi for their help in collecting the nasopharyngeal samples. This research was funded by National Natural Science Foundation of China (No. 81000754), a grant from open subject fund of Jiangsu Provincial Health Bureau of China (No. xk31), a grant from the Key Laboratory for Laboratory Medicine of Jiangsu Province of China (Approval No. XK201114), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and research project funded by the Colleges and Universities in Jiangsu Province plans to graduate research and innovation in 2011.
Disclosure: The authors declare no conflict of interest.
References
- De Lencastre H, Tomasz A. From ecological reservoir to disease: the nasopharynx, day-care centres and drug-resistant clones of Streptococcus pneumoniae. J Antimicrob Chemother 2002;50:75-81.
- García-Rodríguez JA, Fresnadillo Martínez MJ. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother 2002;50:59-73.
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 1995;15:593-600.
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994;264:375-82.
- Martinez-Freijo P, Fluit AC, Schmitz FJ, et al. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother 1998;42:689-96.
- Roy PH. Horizontal transfer of genes in bacteria. Microbiol Today 1999;26:168-70.
- Frost LS, Leplae R, Summers AO, et al. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 2005;3:722-32.
- Mazel D. Integron: agent of bacterial evolution. Nat Rev Microbiol 2006;4:608-20.
- Nandi S, Maurer JJ, Hofacre C, et al. Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci U S A 2004;101:7118-22.
- Kovalevskaia NP. Mobile gene cassettes and DNA integration element. Mol Biol (Mosk) 2002;36:261-7.
- Fluit AC, Schmitz FJ. Resistance integrons and super-integrons. Clin Microbiol Infect 2004;10:272-88.
- White PA, McIver CJ, Deng Y, et al. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol Lett 2000;182:265-9.
- White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother 2001;45:2658-61.
- Wertheim HF, Melles DC, Vos MC, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 2005;5:751-62.
- Liu GY, Tong MQ, Mei Y, et al. Mohnarin 2009 annual report: Bacterial resistance surveillance in patient between 14 and 65 years old. Chin J Clin Pharmacol 2009;27:508-16.
- Valverde A, Coque TM, García-San Miguel L, et al. Complex molecular epidemiology of extended-spectrum beta-lactamases in Klebsiella pneumoniae: a long-term perspective from a single institution in Madrid. J Antimicrob Chemother 2008;61:64-72.
- Ma Y, Yao L, Chen H, et al. Antimicrobial resistance of common bacterial isolates from hospitals in China. Chin J Antibiot 2002;27:129-36.
- Chen Z, Li X, Fan X, et al. Antimicrobial resistance of Staphylococcus aureus. Chin J Infect Chemother 2003;3:13-5.
- Wang F, Zhu D, Wu D, et al. Surveillance of bacterial resistance in Gram-positive cocci. Chin J Infect Chemother 2004;4:1-5.
- Mei Y, Gu B, Wen Y, et al. Surveillance of resistance antibiotic in clinical isolates from First Affiliated Hospital of Nanjing Medical University in 2007. Chin J Antibiot 2009;34:245-50.
- Martin R, Wilox KR. Staphylococcu aureus with reduced susceptibility to vancomycin-US. Morbidity and Mortality weekly reports 1997;46:33.
- Tian S, Chen B. A study on community-associated methicillin-resistant staphylococcus aureus. International Journal of Respiration 2006;26:913-6.
- Fedson DS, Musher DM . Pneumococcal vaccine. In: Plotkin SA, Orenstein WB. eds. Vaccines. 4th ed. Philadelpia, PA: W.B. Saunders Company, 2003:529-88.
- Johnson HL, Deloria-Knoll M, Levine OS, et al. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med 2010;7.pii: e1000348.
- Musher DM. Streptococcus pneumonia. In: Mandell GL, Bennett JE, Dolin R. eds. Mandell, Doulas, and Bennett’s principles and practice of infectious disease. Sixth ed. Philadelphia: Elsevier Inc, 2005; 2392-411.
- Song JH, Lee NY, Ichiyama S, et al. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study. Clin Infect Dis 1999;28:1206-11.
- Scriver SR, Walmsley SL, Kau CL, et al. Determination of antimicrobial susceptibilities of Canadian isolates of Haemophilus influenzae and characterization of their beta-lactamases. Canadian Haemophilus Study Group. Antimicrob Agents Chemother 1994;38:1678-80.
- Tudose C, Bumbăcea D, Bogdan M, et al. Antibiotic resistance of S. pneumoniae and H. influenzae strains isolated from patients with community acquired respiratory tract infections. BACTRO multicenter, multidisciplinary study. Pneumologia 2011;60:30-5.
- Chavanet P, Atale A, Mahy S, et al. Nasopharyngeal carriage, antibiotic susceptibility and serotyping of Streptococcus pneumoniae and Haemophilus influenzae in children attending day care centers. Med Mal Infect 2011;41:307-17.
- Rosser SJ, Young HK. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother 1999;44:11-8.
- Labbate M, Roy Chowdhury P, Stokes HW. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. J Bacteriol 2008;190:5318-27.
- LaPara TM, Burch TR, McNamara PJ, et al. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ Sci Technol 2011;45:9543-9.
- Su HC, Ying GG, Tao R, et al. Occurrence of antibiotic resistance and characterization of resistance genes and integrons in Enterobacteriaceae isolated from integrated fish farms in South China. J Environ Monit 2011;13:3229-36.
- Karczmarczyk M, Walsh C, Slowey R, et al. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. Appl Environ Microbiol 2011;77:7121-7.