Novel hemagglutinin-based influenza virus inhibitors
Introduction
Recent outbreaks of contagious respiratory disease have aroused great concern on potential global pandemic outbreak, which would take toll of millions of human lives. Most of those acute outbreaks are caused by newly emerging viruses, such as highly pathogenic avian influenza virus H5N1 in 1997, bat-origin SARS coronavirus hCoV (SARS-CoV) in 2003, swine-origin influenza virus (pdmH1N1) in 2009, novel SARS-like coronavirus hCoV-EMC in 2012 and 2013, and even the avian-origin influenza virus (H7N9) in 2013 (1). Notably, influenza viruses struck human most frequently, and had already caused at least 20 million deaths in 1918 worldwide (Spanish flu). Wild birds are the natural reservoir of influenza viruses, and they migrate regularly around the world every year, making them the best virus transporter for airborne viral-transmission. An avian strain can adapt to the human host and attain human-to-human transmission capability through acquired mutations. An unexpected human adaptation of an influenza subtype or strain rather than currently circulating influenza viruses may cause pandemic flu.
However, only two classes of anti-influenza drugs are now available in clinic therapy, targeting M2 ion channel and neuraminidase (also named sialidase) expressed on influenza virus envelope, respectively. M2 ion channel inhibitors, including amandatine and rimantadine, block virus uncoating process which is an essential step for the release of viral ribonucleoprotein complexes (vRNPs) into the cytoplasm. Nevertheless, M2 ion channel only exist in influenza A virus, thus adamantines is only effective against influenza A, but not influenza B and C. More importantly, currently circulating viruses in human are mostly resistant to adamantines, which therefore are not recommended for a general and uncontrolled use (2). The second class of anti-influenza drug is neuraminidase inhibitors (NAIs). NAIs inhibit the enzymatic activity of neuraminidase, which is critical for efficient release of progeny viruses from infected cells. Generally used NAIs, oseltamivir and zanamivir, were both approved in 1999 for treatment and preventive use for acute uncomplicated flu caused by influenza A and B. An intravenously administrated NA inhibitor, peramivir, was authorized for emergent treatment of infection with oseltamivir-resistant pdmH1N1 in 2009 by FDA of US, then it was extensively approved for the treatment of infection with influenza A and B in Japan in 2010, and was also emergently approved by Chinese government for the treatment of influenza virus infection under the epidemic of new H7N9 strain in 2013. However, NAI-resistant virus strains are constantly emerging, especially to oseltamivir (3).
Why do the resistant influenza virus strains occur so fast? Because influenza viruses mutate so frequently, they can adapt to most novel mild selection stresses. Influenza viruses possess eight segmented RNA genomes and low proof-reading RNA polymerase, which makes errors with a high frequency in its synthesis of viral RNAs. For example, both emerging influenza virus A/pdmH1N1/2009 and influenza A/pdmH7N9/2013 are reassortment of three different origins or subtypes of influenza A viruses (antigenic shift), and also accumulate many gene mutations (antigenic drift). Taking the emerging viruses A/pdmH7N9/2013 for example (1), they are of avian origin, but only the NA gene is closely related to that from another H7N9 virus (KO14), nevertheless their NA gene is shortened by deletion of five amino acids in stalk region. The HA gene is similar to that of an H7N3 virus (ZJ12) from a nearby region (Zhejiang Province) in China, but possesses Q226L mutation, which may enhance its infectivity in human. All the internal gene segments were closely related to those from avian H9N2 viruses, particularly a virus isolated from a brambling in Beijing. Fortunately, both viruses A/pdmH1N1/2009 and A/pdmH7N9/2013 are mostly sensitive to neuraminidase inhibitors, oseltamivir.
Because influenza viruses mutate so frequently, it is almost impossible to produce a timely and sufficiently effective vaccine to prevent the potential oseltamivir-resistant influenza A viruses epidemic outbreaks. Using anti-influenza agents is still the only way for treatment and prevention at the beginning of the outbreak of a pandemic with a virulent influenza strain, leaving time for the development and widespread dissemination of an effective vaccine.
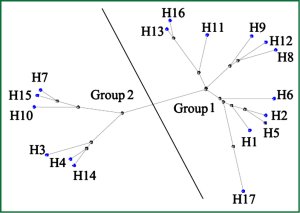
Therefore, it seems quite urgent to seek for novel anti-influenza medications. Up to now, the life cycle of influenza virus has been well understood, allowing the validation of several therapeutic targets. Among them, hemagglutinin (HA) is one of the most appealing targets. The seventeen subtypes of HAs identified can be further subdivided into 2 groups and 5 clades (Figure 1). Notably, H17 HA occurs in the unique influenza virus of bat-origin, which was just reported recently in 2012 (4). This malleable nature of HA imposes a great difficulty to conduct rational drug design, and further, the variety of HA may be even strengthened by antigenic drift and antigenic shift (5,6). Here, we describe the functional and structural studies led to the discovery of HA as new drug target, and also how structural information is facilitating the rational design of novel influenza virus entry inhibitors targeting HA.
Hemagglutinin and hemagglutinin-mediated influenza virus entry
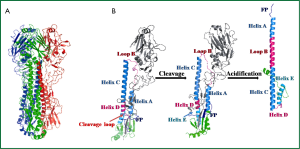
Hemagglutinin (HA), a viral envelope protein, is encoded by the fourth segment of negative-stranded RNA genomes in influenza A or B viruses. This RNA segment initially encodes HA0, the precursor of hemagglutinin. HA0 is post-translationally glycosylated and trimerized with chaperones in endoplasmic reticulum in infected cells. Subsequently, HA0 undergoes an extra- or intra- cellular cleavage process into HA1 and HA2 (Figure 2A,B), which is a critical step for the maturation of influenza virus progenies to acquire their infectivity (7). The HA0 of low pathogenic circulating influenza viruses, with conserved cleavage sites of specified sequence Q/E-X-R, was recognized by extracellular tissue-restrict trypsin-like proteases (8). While for highly pathogenic avian influenza (HPAI) viruses, the HA0 cleavage sites contain multi-basic residues (R-X-R/K-R), and were recognized by ubiquitously expressed intracellular proteases (9).
Through proper cleavage and folding of HA0, three pairs of subunits HA1 and HA2 form fusogenic homotrimer HA (Figure 2A), that is then translocated on the host cell membrane. After viral assembly, progeny viruses bud out by exocytosis, utilizing the cell membrane with HA and NA expression as its envelop. HA protein resembles a “lollipop” on the viral envelop (Figure 2B). The globular head of the “lollipop” is mainly composed of HA1 domains, including receptor binding pocket, vestigial esterase subdomain and antigenic epitopes (10). The receptor binding pocket of HA1 recognizes terminal sialic acids (SAs) of membrane glycoprotein in host cell therefore can prime the virus-cell adsorption and induce sequential endocytosis for virus entry (11). The stem of the “lollipop” is mainly composed of the inner trimric HA2 ectodomain subunits. The first 23 residues of N-terminal of HA2 is the functional fusion peptide (FP), which is accommodated in a hydrophobic pocket formed partially by N- and C-terminal segments of HA1 (12) (Figure 2B). The acidification effect in endosome induces rearrangment of HA1, which facilitates the springing of FP from the inner hydrophobic pocket formed by HA1 (13). Subsequently, low-pH circumstance induces irreversible HA2 reconformation, occurring with FP stretching out and inserting into endosome membrane, and later fusion of the viral envelope with endosome membranes (Figure 2B). The fusion allows the release of viral ribonucleocapsid (vRNP) into cytoplasm of an infected cell, which means viral entry step completed.
Receptor binding site and host tropism
Human adaptation of an avian influenza virus strain rather than currently circulating influenza viruses may cause pandemic flu. However, the change of host tropism from avian to human is mainly dependent on receptor binding preference of influenza virus. The conformation of receptor binding site in HA resembles a shallow “bucket”, consists of β-barrel motif and α-helixes, located on the surface of globular head of HA1 (residues 116-261). Terminal SAs of cell membrane glycoprotein are the natural receptors for viral adsorption, sitting on top of two aromatic residues, Y98 and W153 at the bottom of the “bucket” (12). Besides the bottom, critical binding residues in the “bucket” are conserved amino acids that is edged by 190-helix at the top of the pocket, the 130- and 220-loops located at the front edge and the left side of the pocket, respectively (11). Taking together, the receptor binding sites of HA mainly consists of residues 98, 153 at bottom, 134 to 138 at 130-loop, 183, 190, 194 at 190-helix and 224-228 at 220-loop.
The HA of avian influenza viruses bind predominantly with α-2,3-linked terminal SAs existing mainly in avian digestive tracts. Therefore, avian influenza viruses barely overstep the barrier to infect human which have only α-2,6-linked terminal SAs in upper respiratory tract. In other words, to acquire its infectivity in human, avian influenza viruses have to acquire adequate binding avidity to α-2,6-linked terminal SAs through accumulated antigenic drift or shift.
Consisting with those receptor binding sites reported, several mutations in correspondent binding sites in 130-loop, 190-helix or 220-loop can change HA binding preference for SAs, thus may create a strain of human transmissible avian influenza virus. Recent researches indicate that two mutations in the avian influenza virus H1N1-typed HA1 (E190D in 190-helix and G225D in 220-loop) change the receptor binding preference from the α-2,3-linked SAs to the α-2,6-linked SAs, and those two mutations occur naturally in the 1918 pdmH1N1 and 2009 pdmH1N1 viruses capable of efficient transmission between human without losing their lethality (14,15). Interesting, those two mutations (E190D and G225D) in H5N1 influenza viruses do not cause a switch in receptor-binding specificity (6). Strikingly, in 2012, Imai et al. found that only four mutations (N158D, N224K, Q226L and T318I) in the HA1 of H5N1 are enough for allowing this highly pathogenic virus to be transmitted by respiratory droplets between ferrets (16,17). Notably, novel pdmH9N2 viruses in 2013 also possess a Q226L mutation, which may enhance its infectivity in human (1).
Besides above genetic mutations causing human adaptation of avian influenza, reassortment of the eight segmented genomes of influenza virus can also effectively induce mammal adaptation. By using reverse genetics, Zhang et al. systemically created 127 reassortant viruses between a duck isolate of H5N1, specifically retaining its HA gene throughout, and a highly transmissible, human-infective H1N1 virus (18). Transmission study showed that both polymerase (PA) gene and nonstructural protein (NS) gene of H1N1 virus made the H5N1 virus transmissible by respiratory droplet between guinea pigs, without death. Further experiments indicated that other H1N1 genes are also involved in the enhancement of mammal-to-mammal transmission, including nucleoprotein (NP), neuraminidase (NA), and matrix (M). Hence, avian H5N1 subtype viruses do have the potential to acquire mammalian transmissibility by reassortment in current agricultural scenarios.
Antivirals targeting HA
HA mediates viral entry process, therefore HA inhibitors could block the initial step of viral life cycle, even inducing neutralization of influenza viruses. Obviously, HA inhibitors could stop virus infection immediately, though they may not stop cells infected beforehand from releasing progeny viruses. Applying the strategy of targeting HA, several types of anti-influenza drugs have been discovered or designed, such as antibodies, inhibitors interfering with HA-related factors, peptides, small molecules and even natural small molecules.
Neutralizing antibodies targeting HA
Anti-HA monoclonal antibodies (mAbs) are regarded as one of the important passive therapeutic of influenza. By binding antigenic epitopes of HA, antibodies play a central role in recognition and elimination of invading viruses. Therefore, influenza virus has evolved low-fidelity polymerases that cause frequent mutation and diverse glycosylation of antigenic sites to evade recognition by neutralizing antibodies (19,20). Due to viral antigenic shift and drift, most influenza antibodies and vaccines often expire in a short period. Therefore, it is a leading edge to isolate or design novel broadly neutralizing monoclonal antibodies (bnmAbs), which recognize extremely conserved sites in HA. The highly conserved stem domain of HA2 often elicits cross-reactive antibodies. However, antigenic epitopes in the globular head of HA1 typically elicit strain-specific responses, due to the hyper-variability of this region. Unfortunately, current flu vaccines mainly provoke antibodies that target the head region of HA1, therefore they expire very fast. Otherwise, receptor binding sites of HA1 are more conserved, since any mutations interfering with receptor binding are most likely to impair viral adsorption and infectivity.
Neutralizing antibodies targeting subunit HA1
Based on the fact some antigenic sites overlaps receptor binding sites, these special sites are exploited by naturally occurring antibodies, usually bnmAbs in humans. Antibody F045-092 can neutralize both group 1 and group 2 influenza viruses. F045-092 competes with antibodies that recognize sites antigenic sites in HA1, especially residue S136 in 130-loop, but shows no competition with an antibody (C179) targeting the stem region in HA2 (21). Another two bnmAbs, C05 and CH65, achieve high binding affinities to the receptor binding sites by inserting their heavy-chain CDR3 loop (22,23). Both bnmAbs F045-092 and CH65 were isolated from human B cells, suggesting that receptor-binding site based antibodies occur naturally, which thus are potentially inducible by vaccination strategies (21,23).
Neutralizing antibodies targeting subunit HA2
mAbs, recognizing more conserved and efficacious epitopes in the membrane-proximal stem region of HA2, may broadly inhibit various strains of influenza viruses and prevent emerging of escape mutants. The mAbs binding to fusion peptide (FP) domain were able to inhibit viral fusion and replication in vitro and in vivo (24,25). Recently, Hu et al. obtained a panel of fully human monoclonal antibodies from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient (26). The conserved linear epitope (FIEGGWTGMVDGWYGYHH) targeted by those neutralizing mAbs is part of the FP domain in HA2. Besides antibodies targeting FP domain, mAb CR6261 represents another more important type of HA2-based antibodies. CR6261 was isolated from a person who had been vaccinated against seasonal flu (27). CR6261 exhibits broad neutralizing activity against H1, H2, H5, H6, H8 and H9 influenza subtypes and protects mice from lethal H1N1 or H5N1 virus. Crystal structures of CR6261 Fab in complex with two HAs from lethal H1N1 and H5N1 viruses reveals that this antibody recognizes a highly conserved α-helix (residues 38-58) in the membrane-proximal stem of HA2, thus stabilizes HA2 at the fusogenic pH and blocks viral fusion. However, it is undesired that the conserved epitopes recognized by CR6261-like antibodies mainly stay in the inner surface of HA2 stem region.
Inhibitors interfering with HA-related factors
To infect host cells, progeny viruses must have their HA0s cleaved to form mature functional homotrimer structure, and additionally it is indispensible that sufficient SAs for viral entry have to exist on nearby cell surface. Therefore, elimination of host receptors or inhibition of HA0 cleavage could also block viral entry function.
Virus receptor eliminator: DAS181
DAS181 is a 46 kDa recombinant fusion protein, composed of a sialidase catalytic domain derived from Actinomyces viscosus fused with an epithelial surface-anchoring domain (28). DAS181 is the only host-targeting drug candidate under clinical development in influenza field. Through cleaving virus receptor SAs from the host respiratory epithelial surface, DAS181 extinguishes the essential factor for viral adsorption, and consequently blocks viral entry into target cells. DAS181 cleaves both the α-2,6-SA and the α-2,3-SA, presents a potent inhibitory effect against a panel of laboratory strains and clinical isolates of influenza A and B viruses, including HPAI strains (H5N1), with in vitro EC50 values range from 0.04 to 0.9 nM (28). Recent research revealed that DAS181 is also active against Oseltamivir-resisitant H1N1 strains (29). These findings highlight the potential broad spectrum activity of DAS181 against novel and drug-resistant influenza virus strains.
Inhibitors of HA0 cleavage
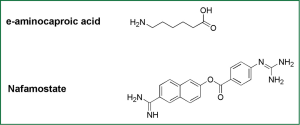
Inhibitors of HA0 hydrolysis interrupt correct folding of HAs in progeny influenza viruses, therefore elicits nonfunctional HA conformation to block viral entry. Some serine protease inhibitors, such as e-aminocaproic acid (8), Nafamostat (30), pulmonary surfactant (a kind of surface active lipoprotein complex) (31) and mucous protease inhibitors (32), can inhibit the cleavage of HA0 and thus viral infection in both cell models and animal models (Figure 3).
As to application of hydrolysis inhibitors in clinic, injection of bovine pancreas trypsin inhibitor, protease peptide (Trasylol bayer), has been applied to treat the symptoms of bleeding. However, it was withdrawn from the clinical application after the study finding that the drug could increase mortality in 2008 (33). Therefore, to avoid severe adverse reaction, natural inhibitors in the human body against HA0 cleavage can serve as a lead compound to develop antiviral drugs which targeting HA. Recently, it was reported the MPSL/TMPRSS13 pathway can hydrolyze H5 and H7 subtypes (34). Inhibitors of MPSL/TMPRSS13 can inhibit the replication of H5 and H7 avian flu with high pathogenicity.
Peptide targeting HA
Peptide drugs are developing very fast in recent two decades. Peptides are small molecules with target specificity and low toxicity. Notably, HA-based peptide vaccine is suggested a promising prophylaxis for influenza. Here, we describes mainly about peptide drugs targeting HA.
Peptides targeting HA1
Obviously, analogs of receptor SA could competitively inhibit viral HA binding with its receptor, therefore inhibit viral adsorption. As alternatives to SAs analogs, using phage-displayed technology, Teruhiko et al. identified several peptides that bind to receptor-binding pocket of H1- and H3- typed HAs, such as N-steroyl peptides C17H35CO-ARLPRTMV-NH2 and C17H35CO-ARLPR-NH2, which can mimic the structure and binding ability of SAs and inhibit the infection of influenza A/H1N1/Puerto Rico/8/34 virus with IC50 values of 3.0 and 1.9 μM, respectively (35). In 2006, a 20-mer peptide (EB), which can specifically bind to the viral HA protein as an entry blocker, was found to exhibit broad-spectrum antiviral activity against influenza viruses in vitro and in vivo, even administered post-infection (36). Laterly, a much shorter peptide P1 (CNDFRSKTC) with similar inhibitory mechanism to EB was also derived (37). These results indicate that the HA-binding peptides are promising candidates as antiviral drugs.
In contrast to HA-binding peptides competitively interfering with HAs on virus envelops, shielding the receptor SAs on host cell membrane is another strategy to block viral adsorption. Alkylation of two peptides c01 and c03 with N-sterol peptides (C18-peptides), derived C18-c01 and C18-c03 respectively. The alkyl groups of the two peptides were able to promote the formation of peptide assembly that ensured multivalent binding with SA-containing receptors. C18-c01 and C18-c03 could inhibit the infection of influenza A/PR/8/34 virus with IC50 values of 3.2 and 6.5 μM, respectively (38).
Peptides targeting HA2
In the early 1990s, several peptides derived from the HIV-1 envelop gp41 HR2 domain potently inhibit HIV fusion and replication. Among those peptides, T20 (generic name: enfuvirtide) was approved by the US FDA as a novel class of anti-HIV drugs known as virus entry inhibitors (39). These discovery enlightened scientists to study possible virus-specific peptides derived from envelope of class I envelope viruses like HIV, such as respiratory syncytial virus (RSV), measles virus, and SARS-CoV.
Since influenza virus also belongs to class I envelop virus, several groups have attempted to identify T20-like anti-influenza peptides. However, to replicate the success of T-20 is not so easy. In contrast to HIV gp41 mediating membrane fusion on outer cell membrane at neutral pH, HA mediates membrane fusion in inner endosome at acidic pH. Therefore, to inhibit fusion process in endosome, active peptide derived from HA2 HR2 region has to permeate cell membrane, enter endosome and stay stable at acidic pH. Up to now, there has been only one study reporting that cholesterol-conjugated HA2-derived peptide inhibits membrane fusion and virus infection (40). They hypothesized that the cholesterol moiety, by localizing the peptide to the target cell membrane, allows the peptide enter the endosome during virus-induced endocytosis. This hypothesis may be advisable for designing fusion-inhibitory peptide derived from HA2.
Small molecules targeting HA
Small molecules occupying small spaces have more possibility to interact with conformational grooves and pockets in the surface of HA protein.
Small molecules targeting HA1
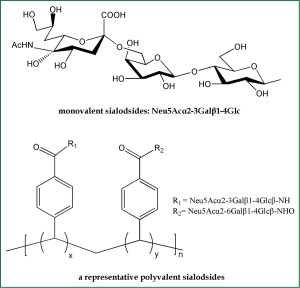
Based on the fact that SA is HA receptors, SA-based inhibitors can be exploited as potential anti-HA agents. Because an influenza virion usually contains about 350-400 HA trimers on its surface (11), monovalent SA analogs would be unable to compete with the highly multivalent interactions between viruses and host cells (Figure 4). Therefore, inhibitors of the bivalent, tetravalent and even polyvalent sialosides afford enhanced inhibitory activity over monovalent ligands (41). The polyvalent sialosides show high potency in vitro (41). It is currently acknowledged that adsorption of α-2,6-linked SAs is indispensable for influenza viruses to gain efficient infectivity in human. Therefore, multivalent α-2,6-sialyloligosaccharides could effectively protect against newly emerging pandemic influenza virus strains.
Small molecules targeting HA2
In the process of HA-mediated fusion, metastable HA2 subunits undergo irreversible rearrangement at acidic pH, which causes membrane fusion and the completion of viral entry. Theoretically, fusion-inhibitory small molecules block influenza virus entry.
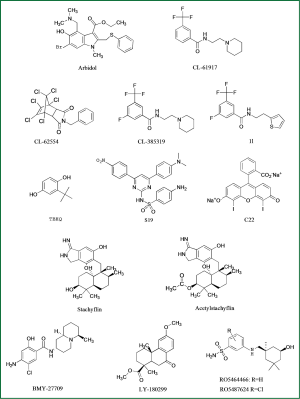
Arbidol is the only influenza entry inhibitor accessible in market, approved only in Russia in 1993 (Figure 5). Arbidol also stabilizes HA2 and prevent acidification-induced reconformation of HA2 (42). A recent study exploiting the characteristics of arbidol-resistant mutants of influenza virus had confirmed its inhibitory mechanism (43), though more mechanisms of action are explained on the inhibitory activity of this magic molecule.
Wyeth-Ayerst researchers discovered three small compounds, CL-61917, CL-385319 and CL-62554, which can inhibit the infection of H1- and H2- subtype influenza viruses with IC50 reaching micromole level (44) (Figure 5). Sequence analysis of drug-resistant mutants of CL-series compounds identified a critical residue mutation in HA2 stem region near FP region. By molecular docking analysis, they revealed that CL-61917 probably binds the cavity near FP domain and may prevent HA2 reconformation triggered by acidification. CL-385319, derived from CL-61917, is more potent than CL-61917 in inhibiting H1 and H2 influenza virus infection.
Our research group further found that CL-385319 also potently inhibits H5N1 influenza virus by targeting a cavity in HA2 stem region near FP domain other than the receptor binding pocket (45). Through site-directed mutation analysis and molecular simulation, we revealed that CL-385319 recognizes and binds with HA in an ‘induced fit’ way, which means that the binding pocket is formed progressively during interactions between CL-385319 and HA (46). By occupying this pocket, CL-385319 stabilizes prefusion conformation of HA2 at fusogenic pH, thus inhibiting the rearrangement required for membrane fusion. Recently, we further optimized the structure of CL-385319, and found that the 3-fluoro-5-(trifluoromethyl)benzamide segment is critical for its anti-H5N1 activity. However, substitution of pyrrolidine ring in CL-385319 with thiophene ring derives compound 1l, which possesses increased anti-influenza activity up to one fold, suggesting 1l is a more attractive candidate with excellent oral bioavailability (47) (Figure 5).
Benzene anthraquinones and hydroquinones can prevent acidification-induced reconformation of HA2. The most promising benzodiazepine derivative is TBHQ, which can inhibit virus infection with IC50 at μM level (48) (Figure 5). TBHQ-resistant mutants revealed that TBHQ mainly interacts with HA2 stem region. Furthermore, cocrystallization analysis of HA combined with TBHQ demonstrated that TBHQ stabilizes the prefusion structure of HA2 at fusogenic pH, mainly by binding with E97 in one HA2 monomer of the HA homotrimer, and R54, E57 in another adjacent HA2 monomer (49). TBHQ can stop the infection of H14-subtype influenza virus rather than H5N1. Through the molecular docking screening, two compounds was found with better antiviral activity, which are S19 (IC50 =0.8 μM) and C22 (IC50 =8 μM) (50), respectively.
Researchers from Shionogi Discovery Research Laboratories in Japan found a few fusion-inhibitory small metabolites, stachyflin and its derivatives, extracted from fermentation broth medium of botryoid ear mould (51) (Figure 5). Stachyflin can effectively prevent the infection of H1 subtype and H2 subtype avian influenza with IC50 reaching a micromole level. Stachyflin stops the viral fusion by blocking HA2 conformational change triggered at low pH. By analyzing the HA sequence of stachyflin-resistant strains, researchers found two point mutations, K51R and K121E located in HA2 subunit (52). This result further confirmed that stachyflin interacts with HA2. The same group also designed and synthesized stachyflin derivatives, such as acetylstachyflin, which have better bioavailability and antiviral activity (53).
Investigators at Bristol Myers Squibb identified a novel influenza virus fusion inhibitor, BMY-27709, which inhibited A/WSN/33 virus replication with IC50 at 3-8 μM and was effective against all H1 and H2 subtype viruses tested (54) (Figure 5). Further sequence analysis of drug-resistant mutants indicated that those compounds prevent HA mediated membrane fusion by interacting with the N-terminal of HA2 subunit (55).
Researchers at Eli Lilly found a derivative of podocarpic acid, LY-180299 inhibits infection of A/Kawasaki/86 (H1N1) strain by affecting an early step of viral replication (56) (Figure 5). Sequence analysis of drug-resistant mutants identified mutations clustered in the interface between HA1 and HA2 and in a region near the FP of HA2. Furthermore, the pH of inactivation of LY-180299 resistant mutants was increased by 0.3-0.6 pH unit, compared with the wild-type viruses. Those data suggest that LY-180299 may interact with the neutral pH conformation of HA, resulting in prevention of the low-pH induced change of HA to its fusogenic conformation.
Recently, Tang et al. reported a series of benzenesulfonamide derivatives with potent anti-influenza activity by targeting HA. These compounds are modified from a salicylamide-based HA inhibitor cis-2-hydroxy-N-(5-hydroxy-1,3,3-trimethyl cyclohexylmethyl)-benzamide (57) (Figure 5). The lead compound RO5464466 and its 2-chloro analogue RO5487624 can effectively prevent the infection of influenza A/Weiss/43 strain (H1N1) with IC50 values of 210 and 86 nM, respectively. In vivo data suggest that RO5487624 potently protected mice challenged with lethal dose of H1N1 influenza viruses (57). Further studies indicated that these compounds inhibit the viral fusion by binding to HA and stabilizing the prefusion structure. These compounds also possess significant metabolic stability, thus could be developed as the first generation HA-inhibitors with excellent oral bioavailability.
Surprisingly, a cheap and widely available anti-malaria drug, chloroquine, may inhibit infection of several viruses including influenza virus in vitro. However, Vigerust et al. found that clinical signs and viral replication were not alleviated by chloroquine in ferrets (58). Just recently, a finished randomised, double-blind, placebo controlled trial also showed that chloroquine does not prevent infection with influenza virus (59). Those results dampen the enthusiasm for potential application of chloroquine for treatment of influenza virus infection. However, researchers at Peking Union Medical College found that anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model if used for therapeutic purpose after infection (60).
Natural products targeting HA
In addition to synthesized or semi-synthesized small molecules, some natural molecules also possess good inhibitory activitiesy against influenza A virus infection (Figure 6). For example, catechins isolated from green tea, like EGCG, were found to exhibit mild anti-influenza effect (61). Further modifications of these catechins derived a set of better inhibitors. Notably, elderberry (Sambucus nigra L.) has been traditionally used for treating influenza and colds in western countries. Highly active flavonoids have been extracted from elderberry, for example, 5,7,3’,4’-tetra-O-methylquercetin achieve an IC50 of 0.36 μM for inhibition of H1N1 infection, comparing favorably to the anti-influenza activities of Oseltamivir (Tamiflu®; 0.32 μM) (62). The direct binding assay indicated that the flavonoids from the elderberry extract bind to H1N1 virions and block viral entry. Additionally, curcumin, the widely used spice and coloring agent in Indian food, was proved to be a good virus entry inhibitor targeting HA with EC50 value approximately 0.47 μM (63). Modification of curcumin may derive a series of novel HA targeting inhibitors. Another remarkable small inhibitor is derived from andrographolide, like AL-1. AL-1 showed significant activity against avian influenza A (H9N2 and H5N1) and human H1N1 influenza A viruses in vitro and in vivo (64). AL-1 was capable of direct interfering with viral HA to block virus binding to cellular receptors, as confirmed by its inhibitory activity on hemaglutination induced by influenza viruses. Therefore, anti-influenza agents from natural products, especially those from traditional Chinese medicine (TCM), are promising lead compounds, which had been extensively reviewed by Xu et al. in 2010 (65).
Conclusions
Hemagglutinin, the major envelope protein of influenza viruses, mediates the viral adsorption, membrane fusion, thus realizing influenza virus entry. The clarified crystal structures of HAs provide resourceful information of the pH-induced rearrangment of HA at the pre-fusion, intermediate and post-fusion states, therefore facilitates the development of influenza entry inhibitors. Although series of anti-influenza drugs targeting the NA and M2 ion channel are currently available in clinic, frequent occurrence of drug-resistance viruses has raised a great concern on potential emerging of a novel influenza virus possessing multi-resistance. Thus, it is essential to develop novel anti-influenza drugs with new targets. Large amount of anti-influenza agents have been shown to block the HA-mediated viral entry by interfering with the receptor binding, the cleavage of HA0, acidic pH-mediated fusogenic rearrangment of HA2, viral entry related factors, and etc. Though there is no full-grown molecular model for discovering or designing small compounds targeting HA1, the clarification of HA2-mediated fusion process provides a great convenience for designing fusion-inhibitory drugs. The promising influenza virus inhibitors may provide more choices for treatment and prevention of severe respiratory infection with drug-resistant viruses and potential pandemic outbreak, besides NA inhibitors and M2 ion channel blockers.
Acknowledgements
This work was financially supported by the National Nature Science Foundation of China (No. 30772602, 81102792). The statements in this paper reflect the reviews of the authors and we apologize for any unintended missed reference in this review.
Disclosure: The authors declare no conflict of interest.
References
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013;368:1888-97. [PubMed]
- Hayden FG, Hay AJ. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr Top Microbiol Immunol 1992;176:119-30. [PubMed]
- Kiso M, Mitamura K, Sakai-Tagawa Y, et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 2004;364:759-65. [PubMed]
- Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 2012;109:4269-74. [PubMed]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981;289:366-73. [PubMed]
- Stevens J, Blixt O, Tumpey TM, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006;312:404-10. [PubMed]
- Jiang S, Li R, Du L, et al. Roles of the hemagglutinin of influenza A virus in viral entry and development of antiviral therapeutics and vaccines. Protein Cell 2010;1:342-54. [PubMed]
- Kido H, Yokogoshi Y, Sakai K, et al. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem 1992;267:13573-9. [PubMed]
- Steinhauer DA. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 1999;258:1-20. [PubMed]
- Sriwilaijaroen N, Suzuki Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci 2012;88:226-49. [PubMed]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 2000;69:531-69. [PubMed]
- Luo M. Influenza virus entry. Adv Exp Med Biol 2012;726:201-21. [PubMed]
- Thoennes S, Li ZN, Lee BJ, et al. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology 2008;370:403-14. [PubMed]
- Tumpey TM, Maines TR, Van Hoeven N, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 2007;315:655-9. [PubMed]
- Maines TR, Jayaraman A, Belser JA, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 2009;325:484-7. [PubMed]
- Auewarakul P, Suptawiwat O, Kongchanagul A, et al. An avian influenza H5N1 virus that binds to a human-type receptor. J Virol 2007;81:9950-5. [PubMed]
- Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012;486:420-8. [PubMed]
- Zhang Y, Zhang Q, Kong H, et al. H5N1 hybrid viruses bearing 2009/h1n1 virus genes transmit in guinea pigs by respiratory droplet. Science 2013;340:1459-63. [PubMed]
- Kim JI, Park MS. N-linked glycosylation in the hemagglutinin of influenza A viruses. Yonsei Med J 2012;53:886-93. [PubMed]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 2008;9:267-76. [PubMed]
- Ohshima N, Iba Y, Kubota-Koketsu R, et al. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 2011;85:11048-57. [PubMed]
- Ekiert DC, Kashyap AK, Steel J, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012;489:526-32. [PubMed]
- Whittle JR, Zhang R, Khurana S, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 2011;108:14216-21. [PubMed]
- Varecková E, Mucha V, Wharton SA, et al. Inhibition of fusion activity of influenza A haemagglutinin mediated by HA2-specific monoclonal antibodies. Arch Virol 2003;148:469-86. [PubMed]
- Gocník M, Fislová T, Sládková T, et al. Antibodies specific to the HA2 glycopolypeptide of influenza A virus haemagglutinin with fusion-inhibition activity contribute to the protection of mice against lethal infection. J Gen Virol 2007;88:951-5. [PubMed]
- Hu W, Chen A, Miao Y, et al. Fully human broadly neutralizing monoclonal antibodies against influenza A viruses generated from the memory B cells of a 2009 pandemic H1N1 influenza vaccine recipient. Virology 2013;435:320-8. [PubMed]
- Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science 2009;324:246-51. [PubMed]
- Malakhov MP, Aschenbrenner LM, Smee DF, et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob Agents Chemother 2006;50:1470-9. [PubMed]
- Triana-Baltzer GB, Gubareva LV, Klimov AI, et al. Inhibition of neuraminidase inhibitor-resistant influenza virus by DAS181, a novel sialidase fusion protein. PLoS One 2009;4:e7838. [PubMed]
- Hosoya M, Matsuyama S, Baba M, et al. Effects of protease inhibitors on replication of various myxoviruses. Antimicrob Agents Chemother 1992;36:1432-6. [PubMed]
- Kido H, Sakai K, Kishino Y, et al. Pulmonary surfactant is a potential endogenous inhibitor of proteolytic activation of Sendai virus and influenza A virus. FEBS Lett 1993;322:115-9. [PubMed]
- Beppu Y, Imamura Y, Tashiro M, et al. Human mucus protease inhibitor in airway fluids is a potential defensive compound against infection with influenza A and Sendai viruses. J Biochem 1997;121:309-16. [PubMed]
- Stamou SC, Reames MK, Skipper E, et al. Aprotinin in cardiac surgery patients: is the risk worth the benefit? Eur J Cardiothorac Surg 2009;36:869-75. [PubMed]
- Okumura Y, Takahashi E, Yano M, et al. Novel type II transmembrane serine proteases, MSPL and TMPRSS13, Proteolytically activate membrane fusion activity of the hemagglutinin of highly pathogenic avian influenza viruses and induce their multicycle replication. J Virol 2010;84:5089-96. [PubMed]
- Matsubara T, Onishi A, Saito T, et al. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J Med Chem 2010;53:4441-9. [PubMed]
- Jones JC, Turpin EA, Bultmann H, et al. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J Virol 2006;80:11960-7. [PubMed]
- Rajik M, Omar AR, Ideris A, et al. A novel peptide inhibits the influenza virus replication by preventing the viral attachment to the host cells. Int J Biol Sci 2009;5:543-8. [PubMed]
- Matsubara T, Sumi M, Kubota H, et al. Inhibition of influenza virus infections by sialylgalactose-binding peptides selected from a phage library. J Med Chem 2009;52:4247-56. [PubMed]
- Kilby JM, Eron JJ. Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med 2003;348:2228-38. [PubMed]
- Lee KK, Pessi A, Gui L, et al. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J Biol Chem 2011;286:42141-9. [PubMed]
- Sun XL. Recent anti-influenza strategies in multivalent sialyloligosaccharides and sialylmimetics approaches. Curr Med Chem 2007;14:2304-13. [PubMed]
- Boriskin YS, Leneva IA, Pécheur EI, et al. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr Med Chem 2008;15:997-1005. [PubMed]
- Leneva IA, Russell RJ, Boriskin YS, et al. Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antiviral Res 2009;81:132-40. [PubMed]
- Plotch SJ, O’Hara B, Morin J, et al. Inhibition of influenza A virus replication by compounds interfering with the fusogenic function of the viral hemagglutinin. J Virol 1999;73:140-51. [PubMed]
- Liu S, Li R, Zhang R, et al. CL-385319 inhibits H5N1 avian influenza A virus infection by blocking viral entry. Eur J Pharmacol 2011;660:460-7. [PubMed]
- Li R, Song D, Zhu Z, et al. An induced pocket for the binding of potent fusion inhibitor CL-385319 with H5N1 influenza virus hemagglutinin. PLoS One 2012;7:e41956. [PubMed]
- Zhu Z, Li R, Xiao G, et al. Design, synthesis and structure-activity relationship of novel inhibitors against H5N1 hemagglutinin-mediated membrane fusion. Eur J Med Chem 2012;57:211-6. [PubMed]
- Bodian DL, Yamasaki RB, Buswell RL, et al. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry 1993;32:2967-78. [PubMed]
- Russell RJ, Kerry PS, Stevens DJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 2008;105:17736-41. [PubMed]
- Hoffman LR, Kuntz ID, White JM. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J Virol 1997;71:8808-20. [PubMed]
- Yoshimoto J, Kakui M, Iwasaki H, et al. Identification of a novel HA conformational change inhibitor of human influenza virus. Arch Virol 1999;144:865-78. [PubMed]
- Yoshimoto J, Kakui M, Iwasaki H, et al. Identification of amino acids of influenza virus HA responsible for resistance to a fusion inhibitor, Stachyflin. Microbiol Immunol 2000;44:677-85. [PubMed]
- Minagawa K, Kouzuki S, Kamigauchi T. Stachyflin and acetylstachyflin, novel anti-influenza A virus substances, produced by Stachybotrys sp. RF-7260. II. Synthesis and preliminary structure-activity relationships of stachyflin derivatives. J Antibiot (Tokyo) 2002;55:165-71. [PubMed]
- Luo G, Colonno R, Krystal M. Characterization of a hemagglutinin-specific inhibitor of influenza A virus. Virology 1996;226:66-76. [PubMed]
- Luo G, Torri A, Harte WE, et al. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J Virol 1997;71:4062-70. [PubMed]
- Staschke KA, Hatch SD, Tang JC, et al. Inhibition of influenza virus hemagglutinin-mediated membrane fusion by a compound related to podocarpic acid. Virology 1998;248:264-74. [PubMed]
- Tang G, Lin X, Qiu Z, et al. Design and Synthesis of Benzenesulfonamide Derivatives as Potent Anti-Influenza Hemagglutinin Inhibitors. Ann Cardiothorac Surg 2011;2:603-7.
- Vigerust DJ, McCullers JA. Chloroquine is effective against influenza A virus in vitro but not in vivo. Influenza Other Respi Viruses 2007;1:189-92. [PubMed]
- Paton NI, Lee L, Xu Y, et al. Chloroquine for influenza prevention: a randomised, double-blind, placebo controlled trial. Lancet Infect Dis 2011;11:677-83. [PubMed]
- Yan Y, Zou Z, Sun Y, et al. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res 2013;23:300-2. [PubMed]
- Song JM, Lee KH, Seong BL. Antiviral effect of catechins in green tea on influenza virus. Antiviral Res 2005;68:66-74. [PubMed]
- Roschek B Jr, Fink RC, McMichael MD, et al. Elderberry flavonoids bind to and prevent H1N1 infection in vitro. Phytochemistry 2009;70:1255-61. [PubMed]
- Chen DY, Shien JH, Tiley L, et al. Curcumin inhibits influenza virus infection and haemagglutination activity. Food Chem 2010;119:1346-51.
- Chen JX, Xue HJ, Ye WC, et al. Activity of andrographolide and its derivatives against influenza virus in vivo and in vitro. Biol Pharm Bull 2009;32:1385-91. [PubMed]
- Ge H, Wang YF, Xu J, et al. Anti-influenza agents from Traditional Chinese Medicine. Nat Prod Rep 2010;27:1758-80. [PubMed]