Critical issues in the clinical application of liquid biopsy in non-small cell lung cancer
Introduction
On the road from the laboratory bench to the patient’s bedside, an assay measuring a tumor marker must be evaluated in order to demonstrate its analytic and clinical validity (respectively: the assay accuracy and reproducibility, and its ability to identify a biologic difference that predicts patient outcome), and at the end its clinical utility (the results of the assay—positive or negative—lead to a clinical decision that evidently improves patient outcome) (1). Thus, a tumor marker must encompass several levels of evidence in order to demonstrate its clinical utility, with the final aim to improve patient outcome, in terms of overall survival (OS), disease-free survival and quality of life, and obtain a more cost-efficient application of effective therapies. The Tumor Marker Utility Grading System (TMUGS) is a framework that establishes an agenda for evaluating the clinical utility of tumor markers, and describes five levels of evidence as “categories that define the quality of data available on which the utility score is based”. These levels range from the weak level V evidence, derived from case reports and clinical examples, to the strongest level I evidence, “the definitive demonstration of clinical utility, obtained by a single, high-powered, prospective, randomized, controlled trial or from a meta-analysis or overview of multiple, well-designed studies”, passing through intermediate degrees of evidence in levels II–IV (2).
Many studies have illustrated the potential of liquid biopsy as tumor marker, to determine the genomic profile of cancer patients, monitor treatment responses and quantify minimal residual disease, and assess the onset of therapy resistance. The concept of liquid biopsy has gradually evolved reaching nowadays a wider field of application from the initial idea of “leukemic phase of solid tumors”, used to indicate the presence of epithelial cells of putative tumor origin in the peripheral blood of cancer patients (3). Circulating tumor cells (CTCs) were described for the first time in 1869 by Ashworth, who documented the presence of cells “identical with those of the cancer itself” in the blood of a metastatic cancer patient (4). In 2001, Pachmann et al. combined laser scanning cytometry with immunomagnetic bead enrichment to detect and quantify rare tumor cells in peripheral blood and bone marrow, describing this method as the most sensitive to “enable extensive investigation of the seeding behaviour of tumours” (5). Three years later, the association of CTC number and patient outcome was demonstrated in metastatic breast cancer using the CellSearch® platform (Janssen Diagnostics, LLC, Raritan, NJ, USA), a semi-automated system that immunomagnetically enriches epithelial cell adhesion molecule (EpCAM)-positive CTCs and enumerates them as (EpCAM+)CK+DAPI+CD45− cells (6).
Since then, substances that could be extracted from blood, serum and plasma have been included in the concept of liquid biopsy, such as circulating cell-free nucleic acids (DNA, RNA or microRNAs), and exosomes (7). Today, the term also incorporates several other biological materials obtained from almost all body fluids (urine, pleural effusion, ascites, etc.) (8).
In the field of CTCs, despite the great number of technologies developed in order to enrich, isolate and characterize these metastatic seeds of tumors, only the CellSearch® technology has demonstrated the clinical validity of CTC quantification (level I evidence) as a prognostic factor—in terms of progression-free survival (PFS) and OS—for metastatic breast cancer, by a pooled analysis of individual patient data published in 2014 (9).
The CellSearch® CTC Kit has been cleared by the Food and Drug Administration (FDA) [2008] for detection and enumeration of CTCs of epithelial origin in whole blood of metastatic colon, prostate and breast cancers. For these pathologies, the EpCAM+ CTC number strongly correlates to prognosis: a cut-off value of CTC/7.5 mL of peripheral blood has been validated, and patients whose CTC number is above this cut-off are at high risk of progression and short OS (6,10-12). Since 2015, the CTC level has been included in the American Society of Clinical Oncology (ASCO) and in the National Comprehensive Cancer Network (NCCN) guidelines for its prognostic value in metastatic breast cancer, consistently with the analyses published by Bidard and colleagues (9). However, its predictive value and clinical utility are still under discussion, and ongoing clinical trials will define the possibility to use this assay for treatment decision together with currently used diagnostic methods.
More in detail, current limits to an extensive use of CTC assays in the clinical setting include: (I) the lack of a consensus about the features that are necessary and sufficient to define an event as CTC; (II) the poor ability to track tumor cells undergoing epithelial-to-mesenchymal-transition (EMT), responsible for the loss of epithelial markers in favor of the mesenchymal ones; (III) the sensitivity of the techniques with a limit of detection of CTCs in no more than 50% of metastatic patients (13); (IV) the lack of a comprehensive genomic characterization of the circulating compartment together with a reliable assessment of its heterogeneity.
In summary, a major limit to the use of CTC assays and, more in general, of liquid biopsy into clinical practice is the lack of a consensus on standard operating procedures (SOPs) for its assessment. Indeed, robust, reproducible and shared procedures are mandatory for reaching the clinical validation of the best assays to isolate and/or characterize the tumor burden in peripheral blood, and for definitively proving their individual or complementary use as companion diagnostic.
This review will summarize current developments on liquid biopsy in non-small cell lung cancer (NSCLC), addressing the limits for the use of CTCs/ctDNA in the clinical practice, and analyzing the solutions adopted to overcome such limits, on the road towards the clinical validation.
Liquid biopsy in lung cancer
Cancer is a leading cause of death worldwide: there were 8.8 million cancer deaths in 2015, most of which due to lung cancer (http://www.who.int/mediacentre/factsheets/fs297/en/). About 80% of lung cancers are classified as NSCLC, 15% are small cell lung cancer (SCLC), and 5% are other histological variants (14). Current therapeutic options are chemotherapy and drug therapy directed against specific molecular targets, mainly epidermal growth factor receptor (EGFR) missense mutations and deletions, and anaplastic lymphoma kinase (ALK) rearrangements. However, response rates are generally modest and patients are still exposed to side effects of both cytotoxic agents and targeted therapy (15). Moreover, most patients are still treated with palliative chemotherapy, due to advanced disease at diagnosis.
Targeted therapies rely on the availability of tumor biopsies for molecular profiling at diagnosis and relapse; these procedures might be invasive, and cause sample bias due to tumor heterogeneity, within the primary tumor and among different metastatic sites, resulting in single-site biopsy being not representative of the complete molecular profile of the systemic disease (16). Thus, current diagnostic tools are hampered by tumor heterogeneity and evolution giving rise to resistance. Furthermore, re-biopsy is not feasible to longitudinally monitor treatment response and resistance development, due to suboptimal quality and poor material recovered by bronchoscopy and invasiveness of the procedure.
Liquid biopsy has emerged as a minimal invasive approach to respond to the urgent need for real time monitoring, stratification, and personalized optimized treatment. In principle, a liquid biopsy could provide the genetic landscape of all cancerous lesions, detecting genomic alterations sensitive to targeted therapy or associated with treatment resistance. Moreover, it would guarantee the prognostic/predictive biomarkers evaluation in patients for whom biopsies are inaccessible or difficult to repeat.
CTCs and circulating tumor microemboli (CTM)
CTCs are cancer cells that detach from the primary tumor or metastatic sites, enter the bloodstream and might develop into further metastases. They are found in around 50% of patients affected by metastatic epithelial tumors (13). CTCs are extremely rare events, occurring at an estimated frequency of one against 106–107 leukocytes (17). They can travel in the bloodstream as single cells or cell clusters, called Circulating Tumor Microemboli (CTM) (18). CTM consist of tumor cells, fibroblasts, leukocytes, endothelial cells, pericytes and platelets held together by the expression of cell adhesion proteins such as plakoglobin. They are considered to play an important role in the development of metastasis, because tumor cells within clusters are protected by both the presence of neighboring cells and the expression of adhesion molecules, and are thus more able to escape from immune surveillance and reach distant organs (19,20). Hou et al. compared the molecular characteristics of CTM and solitary CTCs found in SCLC patients: in CTM, apoptotic cells were absent and tumor cells were not proliferating, supporting the hypothesis that they might have a survival advantage and be more resistant to chemotherapy than individual proliferating CTCs (21).
Cell-free DNA (cfDNA) and circulating tumor DNA (ctDNA)
cfDNA has recently emerged as a promising diagnostic tool for cancer patients: cfDNA originates from both normal and tumor cells that undergo apoptosis or necrosis, and from macrophages that phagocytize necrotic cells. A higher level of cfDNA has been detected in cancer patients when compared to healthy subjects, and in later-stage versus early-stage tumors (16). Notably, cfDNA level can increase also in physiological (intense physical activity) and para-physiological conditions (pregnancy), and daily oscillations are observed due to the circadian rhythm (22). Moreover, specific non-tumor scenarios, such as inflammation, end-stage renal failure, stroke, myocardial infarction, surgery, and trauma are associated with high levels of cfDNA (23).
ctDNA is the cfDNA portion specifically derived from apoptotic, necrotic or living tumor cells that actively release DNA in the circulation. Several studies investigated the association of ctDNA levels and clinical outcome in lung cancer patients. However, since the absolute ctDNA amount is poorly significant as a diagnostic tool, the attention has focused on real-time monitoring of tumor-associated mutations for tracking the response to target therapies and/or the early onset of drug resistance (7). In this context, recent techniques such as digital PCR (dPCR) and next generation sequencing (NGS) have overcome the limit of sensitivity of traditional methods, such as Sanger sequencing, allowing the detection of ctDNA even if it represents a poor fraction of the total cfDNA (<1.0%) (23).
Exosomes
Exosomes are cell-derived vesicles of 40–100 nm in diameter that are released by several cytotypes into the extracellular space. They contain proteins, lipids, DNA, mRNA, microRNAs and other non-coding RNAs, and are thought to be involved in cell communication and metastasis (24,25). There is growing interest towards these nano-sized vesicles because the exosomal molecules are protected from RNase/proteinase-dependent degradation, and can be stably detected in the circulating compartment, making them ideal biomarkers for several clinical applications (26). Interestingly, Taverna et al. isolated plasma exosomes from a chemo naive 70-year-old patient with stage IV NSCLC, harboring an EGFR activating mutation (27). A panel of 8 miRNAs, with a documented role in NSCLC cell lines (28), was analyzed by real-time PCR, and their abnormal expression documented, consistently with previous findings (29). This study revealed that exosomes-derived microRNAs might constitute novel biomarkers in NSCLC diagnosis and prognosis.
Current limits for lung CTC detection
The EpCAM matter—EMT CTCs
Since CTCs are rare and dispersed into a huge amount of blood cells, their detection/isolation usually requires an initial enrichment step in order to increase their concentration. Enrichment methods are based on their biological characteristics (expression of cell surface protein markers, usually EpCAM for positive selection and/or CD45 for negative selection) or physical properties (size, density, deformability, or electric charges). CTCs can then be detected by immunologic, molecular or functional assays. Table 1 reports some of the most commonly used technologies for CTCs enrichment and characterization in lung cancer.
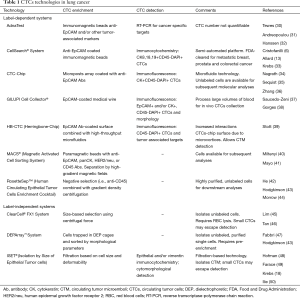
Full table
EpCAM is a membrane glycoprotein highly expressed in the majority of carcinomas, and therefore of potential use as a diagnostic and prognostic marker for a variety of cancers (51). The EpCAM-based technologies allow the detection of CTCs from most epithelial solid tumors. Among those, the CellSearch® system has offered robustness, reproducibility and cost effectiveness, providing the first in vitro diagnostic (IVD) FDA-approved CTC assay for monitoring breast, prostate and colon cancer patients (6,11,12). The system uses ferrofluid particles coated with anti-EpCAM antibodies to capture CTCs. The enriched cells are then stained with fluorescent antibodies anti-panCytokeratin (CK 8, 18, 19; epithelial marker), anti-CD45 (leukocytes), and DAPI for nuclear labeling. A cell is classified as CTC if EpCAM+/CK+/CD45−, with a minimum size of 4 µm × 4 µm, and a DAPI+ nucleus occupying at least 50% of the CK+ cytoplasm. Speaking about NSCLC, the prognostic value of CTCs detected by the CellSearch® system was first reported by Krebs and colleagues in a cohort of 101 patients, with previously untreated stage III/IV NSCLC. Their study demonstrated that patients with 5 or more CTCs/7.5 mL of blood had a worse prognosis compared with those with less than 5 CTCs (33).
Several studies argued that CTCs in NSCLC may be underestimated when using EpCAM-dependent methods, due to EpCAM downregulation during EMT, an important mechanism for tumor invasion and metastasis. This hypothesis was first explored by Farace et al. using in parallel the CellSearch® system and the Isolation by Size of Epithelial Tumor cells (ISET®) technology in 60 patients with metastatic breast (n=20), prostate (n=20) and NSCLC (n=20). ISET® is an EpCAM-independent, filtration-based system, allowing CTCs isolation followed by cytological characterization (Table 1). The study highlighted an important difference in CTC numbers between the two techniques depending mostly on tumor type, and particularly prominent in metastatic NSCLC patients (49). These results were confirmed also by Krebs et al. in 40 chemo-naive advanced NSCLC patients. In this latter study, CTM were detected in 38% of patients with stage III/IV NSCLC by ISET®, whereas they could not be finely characterized by the standard CellSearch® assay, suggesting the need of a customized ad hoc assay for CTM detection with this platform. Furthermore, the analysis of EpCAM expression by immunohistochemistry on ISET®-isolated CTCs/CTM of 9 selected patients showed that all detected cells were negative for this marker, consistently with the lack of CTC detection by CellSearch® in the majority of this subgroup of patients (18).
Even if the EpCAM-independent methods, have demonstrated to detect a higher number of CTCs in NSCLC patients, larger well-designed clinical studies are needed to validate their biological and clinical significance. At this regard, the Foundation for the National Institutes of Health (FNIH) Biomarkers Consortium states that “the mere comparison of two assays metrics/unit blood” is not sufficient to assert that a new assay is more sensitive than a gold standard assay; instead, it is mandatory to determine if the new assay can still retain or even improve the robust capacity of the gold standard to discriminate the outcome of patients who are positive versus those who are negative for the assay (52).
This item was first addressed by De Wit et al. that used an innovative method to investigate the presence of both EpCAM+ and EpCAMlow/– CTCs in the same patient sample, by the collection of the blood discarded by the CellSearch®, after immunomagnetic enrichment of EpCAM+ CTCs, followed by filtration of the EpCAMlow/– cells, and immunofluorescent detection. The study confirmed that the CellSearch® system is very efficient in recovering cells with relatively high EpCAM expression, but less effective with cells showing low/no EpCAM expression. A large portion of cells discarded by the CellSearch® system could be recovered by filtration, however, the efficacy depends on the size of the cells and smaller tumor cells may be missed using this approach. The study confirmed the association between EpCAM+ CTCs and poor patient outcome, whereas no association was found between EpCAMlow/– cells and outcome (53). The small cohort analyzed (27 metastatic lung cancer patients), however, limits the conclusions of this study and highlights the need of a multicenter pooled analysis to get a better picture of the EpCAMlow/– CTCs role in lung cancer.
Concerning EpCAM-negative CTCs and patient outcome, a more recent study from Pailler et al. evaluated the predictive value of ALK-aberrant (rearranged or with copy number gain, CNG) CTC number in 39 metastatic ALK-positive NSCLC patients treated with crizotinib. The group combined immunofluorescence staining (DAPI/CD45) and filter adapted-fluorescence in situ hybridization [FA-FISH; (54)] to perform ALK FISH on ISET®-enriched CTCs (at baseline and at an early treatment time-point). No association was found between baseline ALK-aberrant CTC number and PFS/OS (median ALK-rearranged CTCs 14/3 mL, range 1-119; median ALK-CNG CTCs 12/3 mL, range 1–53). Conversely, during treatment there was a significant association between a decreased ALK-CNG CTC number and longer PFS (median ALK-rearranged CTCs 13/3 mL, range 3–60; median ALK-CNG CTCs 15/3 mL, range 2–177). Although this study highlights the potential use of dynamic changes of ALK-CNG CTC number for monitoring treatment efficacy in ALK-positive NSCLC, its results still apply to a small cohort of patients, and reflect the lack of a consensus on CTC definition (i.e., no epithelial/mesenchymal additional marker for CTC detection by ISET®). Moreover, the parallel CellSearch® analyses revealed a median CTC number of 0/7.5 mL detected both at baseline and during treatment (range 0–713 and 0–544, respectively), thus the EpCAM-positive CTC fraction, that so far has been demonstrated to correlate with NSCLC patient outcome, was not further associated to clinical data in this study (55).
Notably, EMT CTCs and CTM could be only two of the factors affecting CTC detection: in fact, CTCs expressing a different CK-pattern from the most commonly detected in epithelial-dependent assays (CKs 4–6, 8, 10, 13, 18, 19) have been documented in lung cancer (53). Secondly, apoptotic CTCs have been assessed in carcinoma patients, suggesting that the detection of viable cells could be crucial to identify the subset of CTCs most likely involved in the metastatic process (56). All together, these findings underline the urgency of a profound molecular characterization of the different CTC subpopulations identified in NSCLC.
The volume issue: CTC assay sensitivity
Currently, CTCs are detected in around 50% of patients affected by epithelial tumors with distant metastases (13), with the CellSearch® assay identifying 20–40% of CTC positive metastatic NSCLC patients (32,33,48,57). In order to increase CTC detection in several cancer types, lung cancer included, other solutions have been investigated. In particular, recent efforts have focused on sample volume (58) as a crucial technical issue for assays sensitivity, moving towards the processing of large volumes of blood to increase CTC detection, particularly in early stage and/or low CTC rate tumors, such as NSCLC (37).
The Gilupi CellCollector® is a CE-approved device developed in order to screen large volumes of blood (Table 1). The medical device consists of a stainless steel wire with a functionalized surface composed of a hydrogel matrix tip coated with anti-EpCAM antibodies. The device is inserted into the cubital vein of patients for 30 minutes and captures CTCs in vivo. Gilupi CellCollector® obtained promising results on CTCs isolated from several cancer patients with a detection rate of around 70% in both early and late cancer stages (http://www.gilupi.com/cellcollector.html).
Two groups reported the in vivo use of this medical wire in lung cancer: the first study was conducted in 12 NSCLC patients and 29 healthy volunteers. After removal of the device from the patient’s arm vein, bound CTCs were detected by immunofluorescence, as described in Table 1. CTCs were observed in all patients with a median of 16 CTCs (range 2–515), whereas no CTC was detectable in healthy volunteers (37). The second report was a prospective, blinded, single-center clinical study, including 50 patients with newly diagnosed and locally advanced/metastatic lung cancer (48 NSCLC patients) and 10 healthy individuals. Patients underwent two subsequent device applications during the same visit, to assess the reproducibility of measurements with the medical wire, before and after 12 weeks of therapy, and CellSearch® analysis was performed in parallel from 7.5 mL of blood collected immediately before the wire application. CTCs captured by the device were stained and counted similarly to the previous study [(37) and Table 1]. The results of the two subsequent CTC analyses performed in both visits detected >1 CTC in 58% of analyzed wires (median 5 CTCs, range 1–56), with 78% of CTC-positive patients at the first pre-therapeutic visit, and 72% at the second post-therapeutic. The incidence of CTC-positive patients was higher if determined by the wire compared with the CellSearch® technology (27% positive patients, range 1–300 cells), showing a median of 3 more CTCs per patient with the wire. CTC positive wires from 2 patients were further characterized by chip-based dPCR for specific KRAS and EGFR mutations of the primary tumor (38). The advantages of the CellCollector® technology is that the hydrogel matrix can be coated with antibodies directed against different surface markers in order to isolate CTCs with low EpCAM expression, and, moreover, that collection occurs in vivo. A limit, however, is that the volume of blood that gets in contact with the device during the 30 minutes can vary around 1.5–3 L (counts reported as CTC/30 min) (37), probably affecting the quantitative data among different patients or time points in the same patient. Nevertheless, the study from Gorges et al. reported reproducible results between two subsequent device applications during the same visit of the patient, underlining the utility of this method to increase CTC capture rates. Furthermore, they demonstrated the feasibility to perform molecular characterization of device-isolated CTCs that can provide important information on therapeutic targets or resistance mechanisms in cancer patients (38).
Recently, Fischer and colleagues introduced the idea that diagnostic leukapheresis (DLA), could be used to isolate a large number of CTCs even in non-metastatic cancer patients. DLA is a standard method frequently used in the clinical setting to isolate mononuclear cells (MNCs) from peripheral blood for various applications including stem cell harvest. In their study, they suggest that DLA would enable CTCs collection together with MNCs, as they have similar density. CTCs were detected in more than 90% of non-metastatic breast cancer patients by a CellSearch® test adapted to DLA, and a correlation was found between CTC numbers and anatomic disease spread. This study also combined DLA to genomic single-cell profiling and suggested their use in order to improve both the prediction of therapy response and the monitoring of early systemic cancer (59).
High throughput microfluidics: improving CTC recovery
Since CTCs are extremely rare events, occurring at an estimated frequency of one against 106–107 leukocytes (17), the use of high throughput microfluidic-based platforms has emerged in the field of CTC technologies, with the aim to improve their recovery efficiency in the huge amount of contaminating leukocytes. These platforms isolate unlabeled cells—down to a single-cell level—with minimum sample pre-analytical processing, allowing their downstream molecular profiling, ex vivo and in vivo applications (cell cultures, drug testing, xenografts).
The ClearCell® FX system has been recently developed as an EpCAM-independent microfluidic-based enrichment method (Table 1). This technology takes advantage of the microfluidic CTC-Chip (Table 1) to isolate unlabeled CTCs based on size, deformability and inertia. Tan et al. used this system to isolate CTCs from 27 NSCLC patients (12 wild-type, 14 ALK-rearranged and 1 with unknown ALK status) and test them for ALK translocation by FISH, similarly to what Pailler et al. reported in ISET®-isolated CTCs (46,54). The results of both studies provide evidence of highly concordant ALK rearrangement patterns between CTCs and tumors, suggesting a CTC utility as a diagnostic tool, and for monitoring resistance during follow-up.
In the context of ex vivo studies, Zhang et al. implemented a co-culturing model to expand CTCs captured from 19 early stage lung cancer patients. They used a microfluidic device similar to the CTC-Chip (Table 1) to isolate CTCs (68% CTC positive patients, range 1–11/mL, median 3 CTCs), and further expand them in situ, on the chip, setting up an ideal tumor microenvironment (73% expansion efficiency). This ex vivo technique for culturing CTCs opened a new scenario for enriching early stage CTCs, in order to understand their role in the metastatic process (36). Nevertheless, the bias of an in vitro selective pressure that might select the more aggressive CTC clones must be considered.
Exploring the metastatic potential of lung CTCs
Regarding in vivo studies, Hodgkinson et al. demonstrated the tumorigenicity of CTCs enriched from 6 extensive-stage chemotherapy-naive SCLC patients by RosetteSep™ technology (Table 1), and injected in immunocompromised mice. Interestingly, the CTC number detected by CellSearch® in the paired blood samples of those giving rise to CTC-Derived eXplant (CDX) were all >400/7.5 mL, whereas lower CTC numbers did not generate CDX (range 20–1,625; 67% CDX efficiency, 4/6 patients). Moreover, CellSearch®-enriched CTCs from the blood samples of two patients with the highest numbers, giving CDX (1,625 and 1,376 CTCs/7.5 mL, respectively), were recovered using the DEPArray™ system: single CTCs and groups of cells were isolated and genomic profile was performed showing a strong correlation between isolated CTCs and their corresponding CDX, analyzed in parallel (43).
In a more recent case study reported from the same group, the authors obtained a CDX from the blood of a 48-year-old NSCLC patient. Three blood samples were drawn at each visit, before chemotherapy and after completion of brain radiotherapy (after the first cycle of chemotherapy the patient had brain progression), and used for CTC enrichment before mice implantation, CellSearch® and ISET® analyses. A CDX was obtained only from the post-radiotherapy, but not from the baseline sample (4 CTC/7.5 mL). CTC analyses of the post-radiotherapy sample matched to the CDX, showed 0 CTCs/7.5 mL by the CellSearch®, and >150 CTCs/mL by ISET® (CK+ and/or Vimentin+ CD45– CD144–). 77% of CTCs were positive for the mesenchymal marker Vimentin. This study reported for the first time a CDX generated from a NSCLC patient without detectable EpCAM+CK+ CTCs, supporting the idea that not only EpCAM+ (43) but also mesenchymal CTCs have a tumor initiating potential in NSCLC (44). In order to further investigate this hypothesis, it would be mandatory to assess the efficiency of CDXs generated from NSCLC patients with no/few CellSearch® detectable CTCs within larger cohorts of patients.
In these and other in vivo studies, one major limit for an application of xenografts in co-clinical trials focused on direct patient treatment is the large number of CTCs necessary to obtain a CDX, which is hardly detected in NSCLC patients. To date, Rossi et al. obtained mice xenografts from EpCAM-enriched CTCs (CellSearch® Profile Kit) of 5 prostate and 2 breast cancer patients with around 50 CellSearch® detected CTCs in the matched blood sample (2 prostate patients had 51 and 66 CTCs/7.5 mL, respectively; CTC range 51–2,866; 100% engraftment efficiency in peripheral blood, 8/8 mice), thus supporting the role of EpCAM+ CTCs in initiating metastases and opening a new scenario in the understanding of the metastatic potential of CTCs (60).
Genomic characterization of the circulating compartment
NSCLC is one of the most clonal heterogeneous tumors, with a variety of driver-mutations characteristic of different patient subsets. The majority of these genomic alterations include EGFR, ALK, and KRAS genes. Mutations in EGFR gene are commonly localized in the tyrosine kinase domain. Patients whose primitive tumor results positive for EGFR mutations are normally treated with tyrosine kinase inhibitors (TKIs: erlotinib, gefitinib, 1st-generation; afatinib, 2nd-generation) but, unfortunately, often develop resistance within 9–12 months. In many cases, resistance lies on the establishment of secondary mutations such as the EGFR-T790M, responsible for a reduced TKI binding to its ligand (61).
Another target for NSCLC therapy is the EML4-ALK-fusion oncogene, currently assessed on tumor biopsies or fine-needle aspirations. ALK translocation is reported in 2–7% of advanced NSCLC, and is a negative prognostic marker. However, its detection by FISH analyses allows for targeted therapy with crizotinib (1st-generation) and ceritinib (2nd-generation), which is more effective than standard chemotherapy (61).
These tumor “druggable” mutations have been largely investigated in the circulating compartment of NSCLC patients, also thanks to recent highly sensitive methods, such as NGS and dPCR, which allow the detection of low abundance mutations in the context of a huge amount of non-tumor circulating cells/nucleic acids. In principle, the concordance of genomic alterations between tumor biopsies and CTCs/ctDNA has been demonstrated by several reports, both for EGFR mutations (62-64) and ALK translocations (50,54,65), leading to the inclusion of CTCs/ctDNA in clinical trials as prognostic/predictive biomarkers. The results of these studies suggest that monitoring EGFR mutations or EML4-ALK rearrangements in the circulating compartment can reveal therapy resistance also prior to radiological progression evidence, thus guiding the therapeutic decision in the patient clinical course (62,65). In this context, Marchetti and colleagues conducted a feasibility study to analyze EGFR mutations by NGS in CellSearch®-enriched CTCs of 37 locally advanced/metastatic NSCLC patients (stage IIIB/IV) recruited within the TRIGGER study. 15 of the 37 patients analyzed were CTC positive by standard CellSearch® analyses (41% positive patients, range 1–29 cells). Moreover, in 33 cases, “potential neoplastic elements” were detected including cells not fulfilling all CellSearch® criteria (the ‘‘suspicious objects’’ defined by the instrument manual), and isolated or clustered large naked nuclei with irregular shape. All the 37 CellSearch®-derived samples were subjected to mutational analyses by NGS, and results were compared to the mutational status of the primary tumor as detected by Sanger sequencing (all EGFR positive). The study supported the coupling of the CellSearch® system with ultra-deep sequencing as a powerful method for following EGFR mutations in CTCs (31/37 CTC-enriched samples were EGFR positive), and highlighted a 94% concordance of the mutation type between primary tumor and the circulating compartment. Interestingly, in four CTC-enriched samples, the study disclosed multiple EGFR mutations in CTCs by NGS, when the corresponding primary tumor had only a single activating mutation detected by Sanger sequencing. However, the possibility that those multiple mutations could be present also in minor clones of the primary tumor, but being undetectable by Sanger sequencing, could not be excluded because NGS was not performed on the primary tumor (66). Nevertheless, this and other studies document a genetic heterogeneity for EGFR mutations in lung cancer, with cases of multiple EGFR mutations in tumor tissues [detected by NGS, (67)], and CTCs (62). Moreover, a different mutational status between primary tumor and CTCs/ctDNA was also reported elsewhere (32,68). Notably, these studies also indicate the mandatory need to use methods that are more sensitive in the analysis of tumor biopsies at diagnosis, rather than Sanger sequencing.
Tumor heterogeneity might be responsible for progression and pharmacological resistance. In this context, the use of untargeted approaches (array comparative genomic hybridization, whole genome sequencing/exome sequencing) has the advantage, besides the high sensitivity, to allow a wider screening and enable the identification of unknown alterations that might be responsible for a specific cell phenotype (stem-cell, EMT, invasiveness) and/or therapeutic response. In this sense, coupling enrichment and isolation of single CTCs with the most recent developed high-sensitivity assays has revealed a great potential to simultaneously assess tumor heterogeneity and monitor tumor-associated genetic markers (43,47,69).
The long road towards clinical utility
To date, the role of CTCs as a prognostic and predictive biomarker in lung cancer is still puzzling. Several studies associate CTC number to OS and/or PFS of metastatic NSCLC patients (32,33,48,57), meanwhile others do not find these associations (70,71). Many small trials have also demonstrated the association between CTC count and therapy efficacy, comparing the count before and after therapy (38,72-74), or during target therapy, in order to detect the onset of resistance mutations (50,54,62,66,75). Even though most data suggest the utility of CTCs as biomarkers for assessing prognosis and monitoring therapy response in NSCLC, these assays are still not used in routine clinical practice, mainly due to the lack of standardized procedures allowing reliable and reproducible results that could be compared among different cohorts of patients.
This review has addressed the technology issues that contribute to the poor ability to track tumor cells in the blood of NSCLC patients, thus hampering the extensive use of CTCs in the clinical setting. The limit of EpCAM-based CTC technologies, excluding EMT cells from being isolated and quantified, has been discussed. EMT CTCs are documented in NSCLC (76), and low numbers of CTCs are observed with epithelial marker-dependent methods (13,33). Indeed, methods based on physical properties (48,49,53) or PCR-based methods (32), usually detect higher numbers of CTCs in lung cancer. However, so far, the FDA-cleared CellSearch® assay, enriching and counting EpCAM+ CTCs (Table 1) has proved to identify 20-40% of CTC positive metastatic NSCLC patients (32,33,48,57). Moreover, a CTC positive status has been associated with a positive lymph node using this test (32,77). On the other side, the EpCAMlow/− fraction of CTCs alone has not been shown to correlate with patient outcome (53). Consistently, in other types of metastatic tumors, mesenchymal CTCs alone have been only weakly associated with OS and PFS (78). All together, these data suggest that a complementary dual technology, detecting both epithelial and mesenchymal CTCs might allow to obtain a much clear picture of the potential of these cells in predicting patient outcome (18).
Two great issues in CTC detection are assay sensitivity and recovery efficiency. At this regard, we described the processing of large volumes of blood to increase CTC detection, in early non-metastatic patients (59) and in metastatic NSCLC (37,38), conditions in which CTCs occur at very low rate. This approach has revealed the potential to greatly improve CTC sensitivity, allowing recovery of higher cell number for downstream molecular analyses (59), and increasing detection rate up to 70% CTC positive patients in lung cancer (38). In addition, the use of high throughput microfluidic-based platforms has greatly improved CTC recovery in the huge amount of contaminating leukocytes [1 CTC against 106–107 leukocytes (17)], achieving the isolation of pure, unlabeled CTCs (36,43,46). The combination of these technologies allowing rescue of a great number of pure CTCs down to a single cell level, together with the recently developed highly sensitive genomic profile techniques (NGS and dPCR among others), has promising results in terms of personalized management of the NSCLC patients (38).
Similar to CTCs, also in the context of ctDNA, its low abundance, compared to the huge amount of constitutive cfDNA, is a major limit of detection. Also in this case, the availability of highly sensitive tests, has permitted to overcome this issue and deepen analyses of this circulating compartment. Since several other physiological and pathological features can influence its absolute amount, cfDNA is a weak diagnostic tool in the absence of specific tumor-associated mutations (8). For this reason, the clinical interest on ctDNA has focused mainly on real-time monitoring of specific tumor-associated mutations as prognostic biomarker for tracking the response to target therapies and/or the early onset of drug resistance, with promising results in terms of patient follow-up (79).
Together these findings suggest that combined molecular analyses of CTCs and ctDNA in blood samples may be useful to monitor the dynamic changes in the mutation profile that occur during therapy as well as the heterogeneity that emerges as a result of the therapeutic selective pressure. Observations arising from these tumor evolution mechanisms during therapy could be used to define target treatments that suppress the clones responsible of drug resistance before they become clinically relevant.
For all the reasons here described, current studies, focused on reaching a consensus on liquid biopsy utility in NSCLC, are multicenter pooled analyses with larger cohorts of patients, longer observation periods, and CTC/ctDNA molecular characterization according to standardized procedures. In this context, the CANCER ID consortium has started in 2015 (35 partners from 13 countries) with the aim to establish standardized protocols, in order to obtain reliable, reproducible tests, allowing the genomic and molecular characterization of the circulating compartment together with the cross-comparison between different technologies. The final goal is to clinically validate these blood-based biomarkers in NSCLC and breast cancer patients (https://www.cancer-id.eu/).
The long road towards clinical utility of liquid biopsy is probably much harder than it was expected when the “leukemic phase of solid tumors” was first described, but, along the way, the circulating compartment is already revealing its huge potential in the era of precision medicine and personalized care of cancer patients.
Acknowledgements
Funding: The concepts developed in this review were derived from the work performed at the CTC laboratory of IOV-IRCCS, Padova, Italy, and partially funded by Innovative Medicine Initiative Joint Undertaking [115749] CANCER-ID; and Intramural 5X1000 SINERGIA CTC/cfDNA IOV to R Zamarchi. The work of M Manicone is supported by IMI [115749] CANCER-ID. The work of C Poggiana is supported by 5×1000 IOV—Translational Oncology: from benchtop to bedside [DGRV 2980/12 to R Zamarchi].
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med 2009;11:3-14. [Crossref] [PubMed]
- Hayes DF, Bast RC, Desch CE, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst 1996;88:1456-66. [Crossref] [PubMed]
- Mocellin S, Keilholz U, Rossi CR, et al. Circulating tumor cells: the 'leukemic phase' of solid cancers. Trends Mol Med 2006;12:130-9. [Crossref] [PubMed]
- Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J 1869;14:146-9.
- Pachmann K, Heiss P, Demel U, et al. Detection and quantification of small numbers of circulating tumour cells in peripheral blood using laser scanning cytometer (LSC). Clin Chem Lab Med 2001;39:811-7. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Pérez-Callejo D, Romero A, Provencio M, et al. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res 2016;5:455-65. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Bidard FC, Peeters DJ, Fehm T, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol 2014;15:406-14. [Crossref] [PubMed]
- Cristofanilli M, Hayes DF, Budd GT, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol 2005;23:1420-30. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann Oncol 2009;20:1223-9. [Crossref] [PubMed]
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [Crossref] [PubMed]
- Hamilton G, Rath B. Detection of circulating tumor cells in non-small cell lung cancer. J Thorac Dis 2016;8:1024-8. [Crossref] [PubMed]
- Coate LE, John T, Tsao MS, et al. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol 2009;10:1001-10. [Crossref] [PubMed]
- Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, et al. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res 2016;5:466-82. [Crossref] [PubMed]
- Tognela A, Spring KJ, Becker T, et al. Predictive and prognostic value of circulating tumor cell detection in lung cancer: a clinician's perspective. Crit Rev Oncol Hematol 2015;93:90-102. [Crossref] [PubMed]
- Krebs MG, Hou JM, Sloane R, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 2012;7:306-15. [Crossref] [PubMed]
- Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44. [Crossref] [PubMed]
- Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene 2016;35:1216-24. [Crossref] [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Tóth K, Patai ÁV, Kalmár A, et al. Circadian Rhythm of Methylated Septin 9, Cell-Free DNA Amount and Tumor Markers in Colorectal Cancer Patients. Pathol Oncol Res 2017;23:699-706. [Crossref] [PubMed]
- Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015;13:17-24. [Crossref] [PubMed]
- Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015;527:329-35. [Crossref] [PubMed]
- Lin J, Li J, Huang B, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal 2015;2015:657086. [PubMed]
- Taverna S, Giallombardo M, Gil-Bazo I, et al. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget 2016;7:28748-60. [Crossref] [PubMed]
- Garofalo M, Quintavalle C, Di Leva G, et al. MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 2008;27:3845-55. [Crossref] [PubMed]
- Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006;9:189-98. [Crossref] [PubMed]
- Tewes M, Aktas B, Welt A, et al. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat 2009;115:581-90. [Crossref] [PubMed]
- Andreopoulou E, Yang LY, Rangel KM, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int J Cancer 2012;130:1590-7. [Crossref] [PubMed]
- Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. [Crossref] [PubMed]
- Sequist LV, Nagrath S, Toner M, et al. The CTC-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. J Thorac Oncol 2009;4:281-3. [Crossref] [PubMed]
- Zhang Z, Shiratsuchi H, Lin J, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014;5:12383-97. [Crossref] [PubMed]
- Saucedo-Zeni N, Mewes S, Niestroj R, et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol 2012;41:1241-50. [PubMed]
- Gorges TM, Penkalla N, Schalk T, et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin Cancer Res 2016;22:2197-206. [Crossref] [PubMed]
- Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A 2010;107:18392-7. [Crossref] [PubMed]
- Miltenyi S, Müller W, Weichel W, et al. High gradient magnetic cell separation with MACS. Cytometry 1990;11:231-8. [Crossref] [PubMed]
- Mayo C, Ortega FG, Giménez-Capitán A, et al. CK-coated magnetic-based beads as a tool to isolate circulating tumor cells (CTCs) in human tumors. Transl Lung Cancer Res 2013;2:65-71. [PubMed]
- He W, Kularatne SA, Kalli KR, et al. Quantitation of circulating tumor cells in blood samples from ovarian and prostate cancer patients using tumor-specific fluorescent ligands. Int J Cancer 2008;123:1968-73. [Crossref] [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Morrow CJ, Trapani F, Metcalf RL, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol 2016;27:1155-60. [Crossref] [PubMed]
- Lim E, Tay A, Von Der Thusen J, et al. Clinical results of microfluidic antibody-independent peripheral blood circulating tumor cell capture for the diagnosis of lung cancer. J Thorac Cardiovasc Surg 2014;147:1936-8. [Crossref] [PubMed]
- Tan CL, Lim TH, Lim TKh, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [Crossref] [PubMed]
- Fabbri F, Carloni S, Zoli W, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett 2013;335:225-31. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer 2011;105:847-53. [Crossref] [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. [Crossref] [PubMed]
- Trzpis M, McLaughlin PM, de Leij LM, et al. Epithelial cell adhesion molecule: more than a carcinoma marker and adhesion molecule. Am J Pathol 2007;171:386-95. [Crossref] [PubMed]
- Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med 2012;10:138. [Crossref] [PubMed]
- de Wit S, van Dalum G, Lenferink AT, et al. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015;5:12270. [Crossref] [PubMed]
- Pailler E, Adam J, Barthélémy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Pailler E, Oulhen M, Borget I, et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non-Small Cell Lung Cancer Patients. Cancer Res 2017;77:2222-30. [Crossref] [PubMed]
- Rossi E, Basso U, Celadin R, et al. M30 neoepitope expression in epithelial cancer: quantification of apoptosis in circulating tumor cells by CellSearch analysis. Clin Cancer Res 2010;16:5233-43. [Crossref] [PubMed]
- Isobe K, Hata Y, Kobayashi K, et al. Clinical significance of circulating tumor cells and free DNA in non-small cell lung cancer. Anticancer Res 2012;32:3339-44. [PubMed]
- Coumans FA, Ligthart ST, Uhr JW, et al. Challenges in the enumeration and phenotyping of CTC. Clin Cancer Res 2012;18:5711-8. [Crossref] [PubMed]
- Fischer JC, Niederacher D, Topp SA, et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc Natl Acad Sci U S A 2013;110:16580-5. [Crossref] [PubMed]
- Rossi E, Rugge M, Facchinetti A, et al. Retaining the long-survive capacity of Circulating Tumor Cells (CTCs) followed by xeno-transplantation: not only from metastatic cancer of the breast but also of prostate cancer patients. Oncoscience 2013;1:49-56. [Crossref] [PubMed]
- Kumar M, Ernani V, Owonikoko TK. Biomarkers and targeted systemic therapies in advanced non-small cell lung cancer. Mol Aspects Med 2015;45:55-66. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [Crossref] [PubMed]
- Narayan A, Carriero NJ, Gettinger SN, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res 2012;72:3492-8. [Crossref] [PubMed]
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One 2014;9:e103883. [Crossref] [PubMed]
- Marchetti A, Del Grammastro M, Filice G, et al. Complex mutations & subpopulations of deletions at exon 19 of EGFR in NSCLC revealed by next generation sequencing: potential clinical implications. PLoS One 2012;7:e42164. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Park SM, Wong DJ, Ooi CC, et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc Natl Acad Sci U S A 2016;113:E8379-E8386. [Crossref] [PubMed]
- Hirose T, Murata Y, Oki Y, et al. Relationship of circulating tumor cells to the effectiveness of cytotoxic chemotherapy in patients with metastatic non-small-cell lung cancer. Oncol Res 2012;20:131-7. [Crossref] [PubMed]
- Chudasama D, Barr J, Beeson J, et al. Detection of Circulating Tumour Cells and Survival of Patients with Non-small Cell Lung Cancer. Anticancer Res 2017;37:169-73. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Muinelo-Romay L, Vieito M, Abalo A, et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers (Basel) 2014;6:153-65. [Crossref] [PubMed]
- Dorsey JF, Kao GD, MacArthur KM, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non-small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: pilot study results. Cancer 2015;121:139-49. [Crossref] [PubMed]
- Aieta M, Facchinetti A, De Faveri S, et al. Monitoring and Characterization of Circulating Tumor Cells (CTCs) in a Patient With EML4-ALK-Positive Non-Small Cell Lung Cancer (NSCLC). Clin Lung Cancer 2016;17:e173-e177. [Crossref] [PubMed]
- Lecharpentier A, Vielh P, Perez-Moreno P, et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer 2011;105:1338-41. [Crossref] [PubMed]
- Wang J, Wang K, Xu J, et al. Prognostic significance of circulating tumor cells in non-small-cell lung cancer patients: a meta-analysis. PLoS One 2013;8:e78070. [Crossref] [PubMed]
- Bulfoni M, Gerratana L, Del Ben F, et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res 2016;18:30. [Crossref] [PubMed]
- Del Re M, Tiseo M, Bordi P, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget 2017;8:13611-9. [PubMed]


