Lung adenocarcinoma: from molecular basis to genome-guided therapy and immunotherapy
Introduction
Lung cancer is the second most frequently diagnosed tumor in both men and women worldwide, and is the leading cause of cancer-related deaths. In the United States it represents 14% of all neoplasms and is estimated to have produced more than 150,000 deaths in the last year (1,2). This epidemic burden began around the mid-20th century, when the mass-production of packet cigarettes became extended in Western Europe and the United States. Tobacco smoke is the main factor for lung cancer, since it is accepted that it accounts for 80% in males and at least 50% in females. However, although the etiological role of tobacco is crucial, up to 25% of lung cancer presents in people that have never smoked. This is especially evident in women with the adenocarcinoma (ADC) subtype. In these cases, other risk factors such as air pollution, environmental and work related carcinogens also seem to play an important role (3-5). There are two main histological types of lung tumors: small-cell lung cancer (SCLC), and non-small-cell lung cancer (NSCLC). The latter represents 80–85% of these tumors, and different histological subtypes can be distinguished: squamous cell carcinoma (SqCC) (44% in men and 25% in women), pulmonary ADC (28% in men and 42% in women) and large-cell and undifferentiated carcinomas (around 9%); rare subtypes accounting for less than 1% of the cases. The dominant histological type strongly varies depending on the smoking status, ethnic background and geographic location, but nowadays it is accepted that the most frequent is ADC, especially in Asian women (more than 70% in Japanese females) (6,7). Even though the incidence rate of lung cancer has been declining in men since the 1980s and in women since the mid-2000s, and that major efforts have been made in research, smoking prevention, early detection and global healthcare approaches, there have still been no overall significant changes in 5-year survival in the last three decades. Moreover, the 1- and 5-year survival rates in lung cancer are 44% and 17%, respectively, and even in patients with a very early stage disease, when supposedly curative surgery is performed the 5-year survival is less than 60% (2,8).
The use of next-generation sequencing (NGS) technologies has confirmed the prevalence of somatic driver alterations in more than 70% of pulmonary ADC. In fact, the Cancer Genome Atlas (TCGA) has identified that 35% of patients have mutations in oncogene TP53 (tumor suppressor gene 53), overlapping with oncogenic driver alterations such as mutations in KRAS (Kirsten rat sarcoma viral oncogene), EGFR (epidermal growth factor receptor 1 or ErbB1 tyrosine kinase receptor oncogene, also denominated ErbB1 or HER1), BRAF (v-Raf murine sarcoma viral oncogene), MET (mesenchymal-epidermal transition oncogene, encoding a tyrosine kinase receptor), ERBB2 (epidermal growth factor receptor 2 oncogene, also encoding a tyrosine kinase receptor, called ErbB2 or HER2 as well), and fusions in ALK (anaplastic lymphoma kinase), ROS1 (encoding tyrosine-protein kinase ros) or RET (‘rearranged during transfection’, codifying a tyrosine kinase receptor) oncogenes, all of them with potential therapeutic implications. Moreover, in recent years new guided therapies have already appeared that are modifying the prognosis of selected groups of patients who have somatic driver alterations. In contrast with ADC, although TP53 mutations are reported in as much as 81% of SqCC, targetable driver somatic alterations are not frequently found in this tumor subtype (9,10).
Genetic risk factors of lung cancer
As previously mentioned, tobacco smoking is the main risk factor for lung cancer, but there is also an important percentage of never-smokers who develop this tumor. For instance, in the USA, 17,000–26,000 annual deaths can be attributed to lung cancer in never smokers. Thus, environmental carcinogens also seem to play an important role. These external factors appear to combine with genetic susceptibility. In this regard, studies performed both in smokers and never-smokers strongly suggest that polymorphisms in genes involved in DNA repair, cell-cycle regulation, apoptotic pathways, inflammation and telomere length are related with lung cancer (11-14). However, mutation in tumor-suppressor genes also seems to modify susceptibility to this tumor. Both TP53 and TP63 mutations have been reported in patients with either ADC or SqCC. Interestingly, when a tumor-suppressor gene is mutated the risk of multiple neoplasms (including lung cancer) becomes increased. For instance, in the Li-Fraumeni Syndrome, a dominant autosomal disorder, more than half of the affected families have inherited mutations in the TP53 gene and patients present multiple neoplasms in childhood and adolescence. If they survive until adulthood, the risk of tumors, including lung cancer, is highly increased. In turn, TP63 that encodes p63 (tumor suppressor or transformation-related protein 63) is also associated with lung cancer, especially in never-smoker females in Asia (15,16).
Genome-wide association studies (GWAS)
GWAS are population-based studies used to identify single-nucleotide polymorphisms (SNPs) in different genetic loci. The purpose of these genome-wide investigations is to find genetics alleles that are associated with disease phenotypes. At least 28 SNPs have already been observed to be significantly associated with a risk of NSCLC. Of them, three major loci strongly relate to lung cancer: these are 15q25 of the genes encoding neuronal nicotinic acetylcholine receptor (nAChR) (subunit genes CHRNA3 and CHRNB5), 5p15 (TERT and CLPTM1L, genes encoding telomerase reverse transcriptase and cleft lip and palate transmembrane 1-like protein, respectively), and 6p21 (BAT3 or HLA-B associated transcript 3 and MSH5 or MutS Homolog 5 genes, codifying for large proline-rich protein and a MutS protein involved in DNA repair, respectively). These associations are particularly related to lung cancer in specific ethnic groups, such as Caucasians and Asians (17-19)..However, in the vast majority of GWAS, SNPs have demonstrated a strong correlation of polymorphism in two genes, those encoding TERT and CLPTM1L, with lung cancer, indifferently of the ethnic origin of the patients. In particular, TERT polymorphisms are especially associated with ADC in never-smokers. Moreover, GWAS strongly suggest that both TERT and CLPTM1L polymorphisms actually modify the susceptibility to further develop a lung cancer (20-24).
Updated pathological classification of ADC
ADC has become the most common histological subtype of lung cancer in most countries. In 2011 the International Association for the Study of Lung Cancer (IASLC), the American Thoracic Society (ATS) and the European Respiratory Society (ERS) published a proposal of ADC classification that was finally included unchanged in April 2015 in the 4th edition of the WHO Classification of Tumors of the Lung, Pleura, Thymus and Heart (25). Previous editions based the diagnosis of lung cancer on routine histological criteria obtained from resection samples, but the new classification also integrates immunohistochemistry, and gives specific terminology and diagnostic criteria to smaller biopsies and cytology samples. These criteria would be very helpful for clinicians and patients since around 70% of lung cancers are detected now in advanced stages being unresectable. Moreover, patients would be treated with more personalized chemotherapy and/or radiotherapy with the use of the new criteria. Thus, it is very important to differentiate between ADC and other lung tumors, even in small biopsy specimens. Many tumors show clear morphologic features, but if the sample showed no clear squamous or glandular features, a minimal immunohistochemical workup with specific markers would make the difference. At the moment, TTF-1 (thyroid transcription factor 1) and p40 (which recognizes the ΔNp63-a p63 isoform) are the best markers for ADC and SqCC, respectively (6,25-30).
The new ADC classification has interesting innovations. For instance, the term bronchioloalveolar carcinoma (BAC) is no longer used. However, tumors formerly named mucinous BAC are now classified as invasive mucinous ADC, whereas the new name for previously called non-mucinous BAC is lepidic-predominant ADC (25). There is also a new subtype called micropapillary ADC, which has a poorer prognosis. In addition, there are new terms such as AIS (‘in situ’ ADC) and minimally invasive ADC (MIA). Moreover, comprehensive histological subtyping based on the predominant subtype is recommended for invasive lung ADC, and the term “mixed subtype” is not used anymore.
Preinvasive lesions
Atypical adenomatous hyperplasia
This is a small (usually 0.5 cm or even less) atypical proliferation of type II pneumocytes along preexisting alveolar walls, which resembles but falls short of diagnostic criteria for non-mucinous AIS. Atypical adenomatous hyperplasia is most commonly diagnosed as an incidental histologic finding, which is present in 5–20% of lung cancer resection specimens. The appearance of this atypical proliferation in CT scan is the presence of small ground glass nodules of 5 mm or less (25).
In situ ADC (AIS)
This has been considered as a preinvasive lesion in the new ADC classification since it grows purely with a lepidic pattern without invasion. Most of the cases are non-mucinous, with a proliferation of type II pneumocytes or club cells (formerly denominated ‘Clara cells’). More rarely they may be mucinous, with tall columnar goblet cells and abundant mucin in the apical end. The typical image of non-mucinous AIS in the CT scan is to observe small ground glass nodules, whereas the mucinous subtype often has the form of a solid nodule (25). It is worth noting that if AIS is completely resected, the 5-year disease-free survival reaches 100%.
MIA
This concept was introduced to define a relatively benign form of ADC, with nearly a 100% 5-year disease-free survival. MIA refers to a small (≤3 cm) solitary ADC with predominant lepidic growth having an invasion of 5 mm or less. Most of these tumors are non-mucinous, although the mucinous form also exists. Similarly to AIS, while the non-mucinous MIA typically shows ground glass nodes in the CT scan (with a solid component measuring 5 mm or less), the mucinous tumor presents as a solid nodule (25).
Invasive ADC
Invasive ADC is classified according to predominant findings. For this, the use of a comprehensive histological subtyping is mandatory, since it allows the estimation of the percentages of the different components. The latter is currently expressed in a semi quantitative fashion, with 5–10% increments. Tumors of mixed characteristics but containing a predominant lepidic growth pattern of type II pneumocytes and/or club cells (formerly known as non-mucinous BAC), which have an invasive component >5 mm are considered as ‘lepidic predominant ADC’. Moreover, as previously mentioned a micropapillary predominant subtype has been added to the new classification. The signet ring and club cell carcinoma subtypes are characterized by a relatively high percentage of these features. Although the latter are commonly observed in the solid subtype, they can also show acinar or papillary patterns. Interestingly, there is a good correlation between the amount of the ground glass and the solid component in the CT, and the lepidic growth and the invasion of the tissue, respectively (25).
ADC variants
The variants of lung ADC accepted today are invasive mucinous, colloid, fetal and enteric ones. The invasive mucinous ADC (formerly known as mucinous BAC) frequently associates KRAS mutation and lack of TTF-1, and is also characterized by multicentric lung lesions. Histologically, these tumors show different amounts of lepidic, acinar, papillary or micropapillary growth modalities, all of them characterized by the already mentioned columnar cells with abundant apical mucin and small base-oriented nuclei. In this case, the CT scan frequently shows localized or multifocal consolidation, conforming nodules or lobar involvement, as well as air bronchogram (25).
Carcinogenesis and cancer hallmarks
Field change cancerization
Field ‘cancerization’ or ‘effect’ denotes a large variety of loco-regional changes occurring on the surface of tissues that are exposed to carcinogens for a relatively extended period. These cellular and molecular changes, in otherwise apparently healthy cells, predispose to the occurrence of cancerous lesions. The lung, and especially the bronchial epithelium, is a perfect example of field cancerization. A predisposing genetic background along with long-term exposure to tobacco and/or environmental carcinogens, and an appropriate lung tissue microenvironment result in a field susceptibility that could trigger cancer initiation, evolution and progression (31,32).
Epigenetic changes
Epigenetic changes are heritable modifications that affect gene expression and other DNA dependent processes without actually changing DNA sequence (33). Although genetic changes play an essential role in ADC tumorigenesis, epigenetic modifications are also linked to the genesis and progression of cancer, as well as to the response to chemotherapy. These modifications include DNA methylation, and changes in microRNA-mediated regulation and the histone/nucleosome (34). Moreover, different studies have shown a direct association between the presence of methylation of tumor suppression genes and the prognosis of resectable early stage NSCLC. Recently, Daugaard et al. using DNA microarrays, have identified and validated 15 differentially methylated regions (DMRs) in lung ADC, which are absent in the tumor-adjacent normal lung tissue. This study suggests that these DMRs can be used as ADC biomarkers and eventually as targets for novel treatments (35,36).
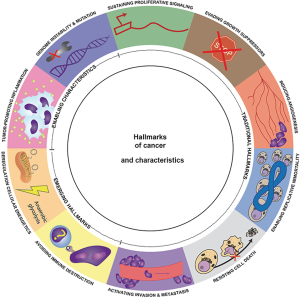
Hallmarks of cancer
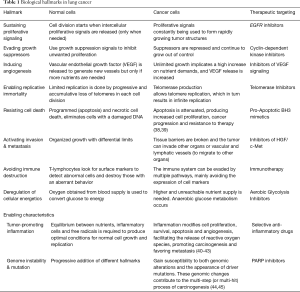
At the beginning of this millennium, Hanahan and Weinberg described the ‘Hallmarks of Cancer’ as the traits that normal cells slowly acquire in their transformation process to a tumor (37). These authors tried to resume the complexity of this process using a multi-hit model, where different characteristics and discrete genetic alterations progressively add up until cancer finally develops. Initially, six hallmarks were described, along with two other emerging findings and two more enabling characteristics that facilitate tumor growth and metastatic dissemination (Figure 1 and Table 1).
Full table
Genomic alterations in lung ADC
As already mentioned, the multi-hit and multi-step cancerogenesis model implies that patients with an intrinsic susceptibility (epigenetic modifications or genome heritable traits) exposed to deleterious factors and with an “appropriate” tumoral-peritumoral environment are predisposed to gain specific somatic genetic alterations (see next section) that trigger an initial clonal cell expansion. At the same time, the aforementioned processes continue to add hallmarks and potentiate an abnormal cell proliferation. This dynamic model conceptualizes cancer as an evolutionary process, where a single cell acquires ‘advantageous’ genomic alterations, allowing itself to proliferate without control, invade and metastasize.
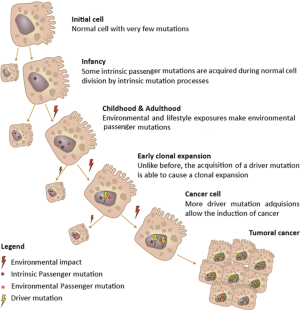
Somatic alterations in cancer genome
Genetic alterations are necessary for oncogenesis. Moreover, all malignant cells show DNA modifications at some point during abnormal proliferation. Although these alterations, which are intrinsic to cancer, can be inherited, most of them are the result of errors when DNA becomes copied during cell cycle. In adulthood, DNA has been copied around 30 trillion times, and a cancer-related mutation can occur at any time, with the probability increasing with the passing of years. These acquired changes in DNA are known as ‘somatic mutations’ or, using a better expression ‘somatic genomic alterations’ (since not all the DNA modifications are mutations). However, not all these changes are related with the development of cancer. Those somatic genomic alterations that are actually involved in carcinogenesis are known as “driver” alterations, whereas those that are not, are called “passenger” alterations (46,47) (Figure 2).
Both pulmonary ADC and SqCC have a high mutational burden compared with other cancers. Interestingly, mutated oncogenes considered as therapeutically targetable predominate in the former. Moreover, when the whole exome of twelve different cancers was sequenced, more than 75% of pulmonary ADC showed driver genomic alterations (48). The frequency of these driver alterations can vary depending on the ethnicity, sex or smoking status, but no differences can be found in different lung ADC stages (49). Table 2 lists the most frequent driver alterations according to TCGA data and the cBioPortal for Cancer Genomics software (open source) (9,50,51).
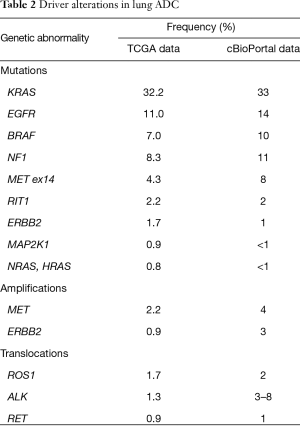
Full table
Epidermal growth factor receptor (EGFR) gene mutations
EGFR is one of the most studied oncogenes related to lung ADC, being located on the short arm of chromosome 7. The EGFR family encodes proteins that belong to the cell-surface tyrosine kinase receptor family, and consists of four members: EGFR (HER1 or ErbB1), HER2 (ErbB2), HER3 (ErbB3) and HER4 (ErbB4) (52-55). These act as transmembrane glycoproteins, and regulate multiple cell processes including apoptosis, cell motility, angiogenesis and proliferative signaling, and also have an impact on carcinogenesis at multiple levels (56,57). EGFR is mutated in 10–16% of ADC, with this percentage being much higher in non-smoking women, especially in Asians (where it reaches a frequency of more than 60%) (58-60). Two different somatic alterations account for more than 90% of the total. One is the L858R mutation (substitution of arginine for leucine at codon 858 in exon 21), which represents 45–50% of the cases, and the other is the E746_A750 deletion (in exon 19) that occurs in 45% of the subjects. In the early stages of the disease, ADC with EGFR somatic alterations has a better prognosis than the “wild-type” tumor after curative resection. Furthermore, even in advanced ADC the presence of EGFR alterations positively changes survival due to the genomic-guided therapy with EGFR tyrosine kinase inhibitors (60-64).
KRAS mutations
KRAS is one of the three members of the so-called RAS family, along with HRAS and NRAS. All of them encode low molecular weight proteins that bind to the Guanosine-Triphosphate (GTP), having crucial roles in monitoring the activity of signaling pathways that control normal cell proliferation (65). Moreover, KRAS mutations were the first somatic alterations that were identified in lung cancer, and despite being a potential therapeutic target, their significance in the clinical setting still remains controversial (66). Besides, they are also the most common mutations detected in lung ADC (33%), being more frequently detected in older men, smokers, and in large-sized solid tumors and poorly differentiated ADC (67-69). Mutations in codon 12 are the most frequently detected (75% of the total) and result in the substitution of glycine for cytosine (Gly12Cys), valine (Gly12Val) or aspartic acid (Gly12Asp), meanwhile mutations in codon 13 are much less observed (around 7%). Unlike EGFR mutations, those occurring in KRAS are strongly related with a poorer prognosis in both early stages of ADC and advanced disease. Unfortunately, the attempts to use guided-therapies to target this mutation-phenotype have been extraordinarily frustrating up to now (67,70-73).
BRAF mutations
BRAF encodes a protein called B-Raf that constitutes a crucial step in the RAS-mitogen activated protein kinase (RAS-MAPK) signal pathway. BRAF mutations are present in 7–10% of patients with pulmonary ADC, and the vast majority of these mutations are characterized by the substitution of valine by glutamate (Val600Glu or V600E) in exon 15 (74,75). Compared with other lung cancers, BRAF mutations are almost exclusive to ADC, although their frequency is low compared with that in other extrathoracic cancers such as melanoma (50–66%) and colorectal carcinoma (>15%). Moreover, this driver mutation is more likely to be observed in smokers and women, and can be targeted by B-Raf protein inhibitors (previously experienced in other cancers). Unlike EGFR or KRAS alterations, the presence of BRAF mutations are not associated with changes in prognosis (65,76,77).
Neurofibromin gene (NF1) mutations
NF1 is an oncogene encoding the neurofibromin protein. This gene is located in chromosome 17 and is composed by 60 exons, making it one of the largest genes in the human genome. This oncogene has been widely described in the context of type 1 neurofibromatosis, and acts as a tumor suppressor with a negative-regulation of the RAS oncogene (78,79). Neurofibromin also regulates cell adhesion, migration and survival, producing a proapoptotic effect. Patients with neurofibromatosis type 1 are considered at high risk of developing malignancies. It should be noted that since TCGA data of somatic mutations are available, NF1 mutation has become a potential therapeutic target both in ADC and SqCC. Patients with lung cancer and NF1 mutation have a concomitant mutation in KRAS in 15% of the cases, but in around 70% exhibit no other somatic alteration. It is worth noting that patients with NF1 alterations in the tumor and those with KRAS abnormalities share similar clinical characteristics and prognosis (80).
MET amplifications and mutations
MET is an oncogene that encodes for the transmembrane MET tyrosine receptor kinase, with only one known ligand (the hepatocyte growth factor or HGF). The presence of MET alterations has a negative impact on prognosis, since amplifications of this gene are related with resistance to EGFR-guided therapy in patients with advanced disease, and a high MET oncogene copy number is associated with worse prognosis in patients with localized disease. However, MET mutations (mutually exclusive with those occurring in KRAS-EGFR), despite being identified with a relative high frequency in ADC, have not been related with an oncogenic potential (54,81-84).
ALK translocations
The ALK gene is located on chromosome 2 and encodes a transmembrane tyrosine kinase. Nearly 30 different ALK fusions have been described, including the EML4-ALK fusion, which is frequently observed in lung ADC (85). This fusion is created by an inversion of the short arm of chromosome 2 that binds exons 1–13 of EML4 (encoding echinoderm microtubule associated protein like 4) to exons 20–29 of ALK, resulting in the synthesis of a chimerical protein with constitutive ALK activity (86-88). Patients with ALK-rearranged ADC are usually young, never-smokers and women, showing moderately or poorly differentiated peripheral tumors (89,90). In general, ALK alterations are mutually exclusive with KRAS-EGFR mutations, having prognosis implications due to the impact of guided-therapies (91).
ROS1 translocations
ROS1 is an oncogene that encodes tyrosine kinase receptor, being phylogenetically related to ALK. Unlike ALK translocations, ROS1 rearrangements include one of twelve different partner proteins, and in lung ADC its fusion with CD74 (cluster of differentiation74), EZR (codifying protein ezrin), SLC24A2 (encoding the sodium/potassium/calcium exchanger 4) or FIG (encoding the fused in glioblastoma protein) genes has emerged as a new driver alteration with promising therapeutic implications. In NSCLC patients the presence of a ROS1-rearrangement is specific for ADC, being frequently observed in Asiatic young women and never-smokers (92-94).
Molecular profiling in lung ADC: when and how?
The complete genetic profile of lung ADC is not easily available in standard clinical practice due to the needs of relatively large tissue samples, which often involve the use of invasive techniques, as well as a good molecular biology laboratory, with properly trained personnel, and the elevated costs of the procedure. For these reasons, the realization of strongly directed molecular tests, aimed at the identification of genetic markers with clinical implications is recommended. In this regard, a useful genetic marker should: (I) be implicated in the tumorigenesis (such as driver alterations) because the pathway suppression could control tumor proliferation; (II) have a high prevalence, to justify the benefit of a costly test; (III) have a highly sensitive and specific validated test; and (IV) have a previously designated oncogenic pathway, with an already available targeted therapy. Although some years ago, a panel of experts from IASLC, ATS and ERS recommended molecular testing only for the EGFR mutation in advanced ADC, more recent recommendations also include EML4-ALK rearrangement in advanced-stages of lung ADC (either locally advanced or metastatic cancer) (95). However, the latest advances in molecular profiling and guided therapies strongly suggest that the screening should already be extended to at least detection of ROS1 fusions, BRAF mutations and MET amplifications or exon 14 alterations, performing a wider genomic profiling in any stage of ADC (26,64,96,97). This will give a more precise scenario of the phenotype epidemiology of this cancer, acting as a strong stimulus for oriented translational research (98).
The first step for the entire process is to identify the origin of the tumor using immunohistochemical techniques in the available sample. Then, the genetic profile is obtained through different techniques such as fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR) or immunohistochemistry. For this, surgical or core-needle samples are preferred due to their larger size. However, molecular techniques can also be applied in smaller samples, such as those obtained in non-invasive or semi-invasive procedures. In this regard, multiple studies have confirmed the utility of even cytological samples obtained by endobronchial ultrasound (EBUS) to perform the molecular study (99-102). However, although a cell block can be obtained by EBUS in most cases (103), there is still controversy on its advantages and disadvantages with respect to the on-site smear in identifying driver alterations (104,105).
Genome-guided therapy
In November 2004, the first genome targeted therapy was approved by the FDA for the treatment of NSCLC with EGFR mutations. Since then, the prognosis of selected patients with advanced ADC and driver mutations has improved substantially. In fact, molecular testing is performed routinely in locally advanced or metastatic ADC since targeted therapies have been approved and their impact on multiple outcomes has been demonstrated. This is the case of patients with EGFR mutations, EML4-ALK rearrangement or ROS1 fusions (64). For instance, ertenolib, gefitinib and afatinib are used in the treatment of locally advanced or metastatic tumors with EGFR exon 19 deletion or exon 21 mutations, while osimertinib, olmutinib and osimertinib are employed in the case of EGFR T790M mutations (106-112). Crizotinib, ceritinib and alectinib in turn are used in similar tumors, which in this case show ALK alterations. If a ROS1 translocation is present, crizotinib can be used to treat the patients (93,113-117). More recently, promising evidence has been published with the use of crizotinib in tumors with MET exon 14 alterations or amplification, and dabrafenib plus trametinib in patients with BRAF mutations (97,118). Table 3 summarizes the approved genome-guided therapies for lung ADC and their present indications.
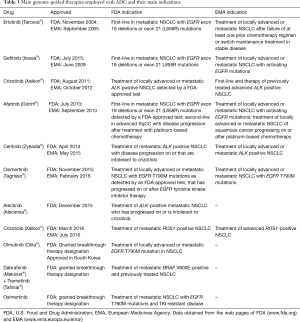
Full table
Immunotherapy
Immunotherapy is a relatively novel approach for cancer, being based on the stimulation of the patient’s immune system to induce a cellular-humoral response that attacks and destroys the malignant cells. Immunotherapy can be active or passive, with both being specific or non-specific. The active immunotherapy consists in the activation of the host’s immune system to induce a specific response, whereas passive immunotherapy is based on the administration of antibodies that will directly kill cancer cells, without interacting with the patient´s immune system. Therapy is specific if it results in a particular immune response or as general if it involves a wider immunological reaction (119).
Many scientific advances in cancer treatment are being developed in the field of active immunotherapies, whose main modalities are therapeutic vaccines and checkpoint inhibitors (120,121). The former stimulates the host immune system to generate a prolonged immunological response by recognizing tumor antigens. The vaccines can be antigen-specific or addressed to the whole-tumor, and have already been studied in the adjuvant setting, as first line and maintenance treatments, but unfortunately no positive results have been found up to now (122-124). Immune checkpoints, in turn, are inhibitory trails that control the duration and intensity of the immune response to reduce the damage in normal tissues. There are two targetable checkpoints that have been widely studied in the last years: the cytotoxic T-lymphocyte antigen 4 (CTLA-4) and the programmed death-ligand 1/programmed cell death protein 1 (PD-L1/PD-1) pathway (125).
CTLA-4 inhibitors
Two humanized monoclonal antibodies inhibiting CTLA-4 have been tested in clinical trials on patients with NSCLC cancer. In this respect, a trial using tremelimumab in advanced-stage NSCLC showed a good tolerability profile but unfortunately showed no differences in the progression-free survival when used as a second-line agent if compared with the best supportive care (126). Two other clinical trials (ClinicalTrials.gov, numbers NCT02000947–NCT02352948), that are now in the recruitment phase, have been designed to compare dual checkpoint inhibition (anti PD-L1 and CTLA-4) using tremelimumab and durvalumab with the standard therapy (127,128).
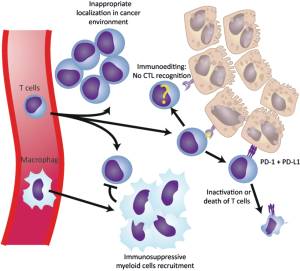
PD-1/PD-L1 inhibitors
Under normal conditions, the PD-1 protein checkpoint protects against inflammation and autoimmunity. When a neoplasm occurs, PD-1 binds to the PD1-L1 and causes immunosuppression, preventing the immune system from attacking the tumoral cells (Figure 3) (129). To date, FDA has approved three PD-1/PD-L1 inhibitor drugs for the treatment of advanced-stages of NSCLC. These are nivolumab (Opdivo©, October 2015), pembrolizumab and atezolizumab (Keytruda© and Tecentriq©, respectively, both in October 2016). Nivolumab is an IgG4 monoclonal antibody that blocks PD-1 receptors expressed on activated T cells. Multiple clinical trials (CheckMate trials) have evaluated nivolumab versus docetaxel in advanced-stage NSCLC, showing an overall improved survival and a significantly better progression-free survival in the nivolumab group, with an acceptable tolerability and toxicity profile, turning this treatment into the second-line gold standard therapy in such cases (130,131). Pembrolizumab, previously called lambrolizumab, is a humanized IgG4 immunoglobulin with a high affinity for PD-1. Many clinical trials (KEYNOTE trials) have shown benefits in the overall response rate (ORR), and the overall survival in a large number of patients with advanced-stage NSCLC when compared with standard therapies, again with an excellent security profile (132,133). Ongoing studies are trying to define if pembrolizumab can be used as a first-line treatment in advanced NSCLC. Finally, a randomized, phase 3 clinical trial (OAK study), with more than a thousand patients from 31 different countries, has shown a better overall survival in patients with a previously treated NSCLC with atezolizumab when compared to docetaxel, irrespectively of PD-L1 expression (134).
In conclusion, the use of genomic phenotyping of ADC, possible now even in relatively small samples, facilitates a better tumor classification, and allows for a more targeted treatment. For this, two different strategies have been developed, genome-guided therapies, mainly based on blocking the aberrant resultant pathway, and immunotherapy, which can either be active (stimulation of the patient's immune system to produce a specific response) or passive (administration of external antibodies). Although the immune strategy is still being developed, its current results are very promising.
Acknowledgements
To Jonathan McFarland for his editing aid.
Funding: This work was funded by FUCAP Grant Agusti Vidal 2015, SEPAR Grant 2015, GSR 2014SGR424 and CIBERES (ISCIII).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- American Cancer Society. Cancer Facts & Figures, Annual report 2016:1-9. Acceded (May 13th 2017). Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2016.html
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Gibelin C, Couraud S. Somatic alterations in lung cancer: Do environmental factors matter? Lung Cancer 2016;100:45-52. [Crossref] [PubMed]
- Ruano-Ravina A, Fernández-Villar A, Barros-Dios JM. Residential Radon and Risk of Lung Cancer in Never-Smokers. Arch Bronconeumol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Travis WD, Brambilla E, Müller-Hermelink HK et al. editors. World Health Organization classification of tumours: Tumours of lung, pleura, thymus and heart. Geneva: WHO Press, 2004:9-122.
- Sanchez-Salcedo P, Berto J, de-Torres JP, et al. Lung cancer screening: fourteen year experience of the Pamplona early detection program (P-IELCAP). Arch Bronconeumol 2015;51:169-76. [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM Classification for lung cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Kim HS, Mitsudomi T, Soo RA, et al. Personalized therapy on the horizon for squamous cell carcinoma of the lung. Lung Cancer 2013;80:249-55. [Crossref] [PubMed]
- Rivera GA, Wakelee H. Lung Cancer in Never Smokers. Adv Exp Med Biol 2016;893:43-57. [Crossref] [PubMed]
- Wu X, Amos CI, Zhu Y, et al. Telomere dysfunction: a potential cancer predisposition factor. J Natl Cancer Inst 2003;95:1211-8. [Crossref] [PubMed]
- Wenzlaff AS, Cote ML, Bock CH, et al. CYP1A1 and CYP1B1 polymorphisms and risk of lung cancer among never smokers: a population-based study. Carcinogenesis 2005;26:2207-12. [Crossref] [PubMed]
- Park JY, Park JM, Jang JS, et al. Caspase 9 promoter polymorphisms and risk of primary lung cancer. Hum Mol Genet 2006;15:1963-71. [Crossref] [PubMed]
- Kleihues P, Schäuble B, zur Hausen A, et al. Tumors associated with p53 germline mutations: a synopsis of 91 families. Am J Pathol 1997;150:1-13. [PubMed]
- Okazaki I, Ishikawa S, Sohara Y. Genes associated with succeptibility to lung adenocarcinoma among never smokers suggest the mechanism of disease. Anticancer Res 2014;34:5229-40. [PubMed]
- Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40:616-22. [Crossref] [PubMed]
- Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 2008;40:1407-9. [Crossref] [PubMed]
- Yang IA, Holloway JW, Fong KM. Genetic susceptibility to lung cancer and co-morbidities. J Thorac Dis 2013;5:S454-62. [PubMed]
- Wang HM, Zhang XY, Jin B. TERT genetic polymorphism rs2736100 was associated with lung cancer: a meta-analysis based on 14,492 subjects. Genet Test Mol Biomarkers 2013;17:937-41. [Crossref] [PubMed]
- Li T, Xian Y, Tian T, et al. New evidence of TERT rs2736098 polymorphism and cancer risk: an updated meta-analysis. J Buon 2016;21:491-7. [PubMed]
- Campa D, Rizzato C, Stolzenberg-Solomon R, et al. TERT gene harbors multiple variants associated with pancreatic cancer susceptibility. Int J Cancer 2015;137:2175-83. [Crossref] [PubMed]
- Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 2009;41:899-904. [Crossref] [PubMed]
- Zienolddiny S, Skaug V, Landvik NE, et al. The TERT-CLPTM1L lung cancer susceptibility variant associates with higher DNA adduct formation in the lung. Carcinogenesis 2009;30:1368-71. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. editors. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. Fourth edition. Geneva: WHO Press, 2015.
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Travis WD. Classification of lung cancer. Semin Roentgenol 2011;46:178-86. [Crossref] [PubMed]
- Travis WD. The 2015 WHO classification of lung tumors. Pathologe 2014;35:188. [Crossref] [PubMed]
- Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med 2011;32:22-31. [Crossref] [PubMed]
- Cagle PT, Allen TC, Bernicker EH, et al. Impact of recent developments in lung cancer on the practice of pathology. Arch Pathol Lab Med 2016;140:322-5. [Crossref] [PubMed]
- Lochhead P, Chan AT, Nishihara R, et al. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol 2015;28:14-29. [Crossref] [PubMed]
- Lee JJ, Liu D, Lee JS, et al. Long-term impact of smoking on lung epithelial proliferation in current and former smokers. J Natl Cancer Inst 2001;93:1081-8. [Crossref] [PubMed]
- Holliday R. The inheritance of epigenetic defects. Science 1987;238:163-70. [Crossref] [PubMed]
- Barreiro E, Gea J. Epigenetics and muscle dysfunction in chronic obstructive pulmonary disease. Transl Res 2015;165:61-73. [Crossref] [PubMed]
- Daugaard I, Dominguez D, Kjeldsen TE, et al. Identification and validation of candidate epigenetic biomarkers in lung adenocarcinoma. Sci Rep 2016;6:35807. [Crossref] [PubMed]
- Ansari J, Shackelford RE, El-Osta H. Epigenetics in non-small cell lung cancer: from basics to therapeutics. Transl Lung Cancer Res 2016;5:155-71. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol 2007;35:495-516. [Crossref] [PubMed]
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007;26:1324-37. [Crossref] [PubMed]
- DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev 2010;29:309-16. [Crossref] [PubMed]
- Grivennikov SI, Greten FR, Karin M. Immunity, Inflammation, and Cancer. Cell 2010;140:883-99. [Crossref] [PubMed]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39-51. [Crossref] [PubMed]
- Karnoub AE, Weinberg RA. Chemokine networks and breast cancer metastasis. Breast Dis 2006-2007;26:75-85. [Crossref] [PubMed]
- Berdasco M, Esteller M. Aberrant Epigenetic Landscape in Cancer: How Cellular Identity Goes Awry. Dev Cell 2010;19:698-711. [Crossref] [PubMed]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007;128:683-92. [Crossref] [PubMed]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719-24. [Crossref] [PubMed]
- Gaughran SJ, Pless E, Stearns SC. How elephants beat cancer. Elife 2016;5:e21864. [Crossref] [PubMed]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep 2013;3:2650. [Crossref] [PubMed]
- Chalela R, Bellosillo B, Curull V, et al. Prevalence of molecular changes in resected pulmonary adenocarcinomas. Eur Respir J 2016;48:PA2853.
- Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Tsiambas E, Lefas AY, Georgiannos SN, et al. EGFR gene deregulation mechanisms in lung adenocarcinoma: A molecular review. Pathol - Res Pract 2016;212:672-7. [Crossref] [PubMed]
- Antonicelli A, Cafarotti S, Indini A, et al. Egfr-targeted therapy for non-small cell lun cancer: Focus on EGFR oncogenic mutation. Int J Med Sci 2013;10:320-30. [Crossref] [PubMed]
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. [Crossref] [PubMed]
- Sharma S V, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2-16. [Crossref] [PubMed]
- Cheng L, Zhang S, Alexander R, et al. The landscape of EGFR pathways and personalized management of non-small-cell lung cancer. Future Oncol 2011;7:519-41. [Crossref] [PubMed]
- Ha SY, Choi SJ, Cho JH, et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget 2015;6:5465-74. [Crossref] [PubMed]
- Tomita M, Ayabe T, Chosa E, et al. Epidermal growth factor receptor mutations in Japanese men with lung adenocarcinomas. Asian Pac J Cancer Prev 2014;15:10627-30. [Crossref] [PubMed]
- Nishii T, Yokose T, Miyagi Y, et al. Prognostic value of EGFR mutations in surgically resected pathological stage I lung adenocarcinoma. Asia Pac J Clin Oncol 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Soria JC, Mok TS, Cappuzzo F, et al. EGFR-mutated oncogene-addicted non-small cell lung cancer: Current trends and future prospects. Cancer Treat Rev 2012;38:416-30. [Crossref] [PubMed]
- Zhu JQ, Zhong WZ, Zhang GC, et al. Better survival with EGFR exon 19 than exon 21 mutations in gefitinib-treated non-small cell lung cancer patients is due to differential inhibition of downstream signals. Cancer Lett 2008;265:307-17. [Crossref] [PubMed]
- Hirsch FR, Suda K, Wiens J, et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388:1012-24. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Santos E, Martin-Zanca D, Reddy EP, et al. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science 1984;223:661-4. [Crossref] [PubMed]
- Lee B, Lee T, Lee S, et al. Clinicopathologic characteristiscs of EGFR, KRAS, and ALK alterations in 6,595 lung cancers. Oncotarget 2016;7:23874-84. [Crossref] [PubMed]
- Dacic S, Shuai Y, Yousem S, et al. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol 2010;23:159-68. [Crossref] [PubMed]
- Rekhtman N, Ang DC, Riely GJ, et al. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol 2013;26:1307-19. [Crossref] [PubMed]
- Kadota K, Sima CS, Arcila ME, et al. KRAS Mutation Is a Significant Prognostic Factor in Early-stage Lung Adenocarcinoma. Am J Surg Pathol 2016;40:1579-90. [Crossref] [PubMed]
- Califano R, Landi L, Cappuzzo F. Prognostic and Predictive Value of K-RAS Mutations in Non-Small Cell Lung Cancer. Drugs 2012;72:28-36. [Crossref] [PubMed]
- Meng D, Yuan M, Li X, et al. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: A systematic review with meta-analysis. Lung Cancer 2013;81:1-10. [Crossref] [PubMed]
- Nadal E, Chen G, Prensner JR, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol 2014;9:1513-22. [Crossref] [PubMed]
- Gautschi O, Milia J, Cabarrou B, et al. Targeted Therapy for Patients with BRAF-Mutant Lung Cancer Results from the European EURAF Cohort. J Thorac Oncol 2015;10:1451-7. [Crossref] [PubMed]
- Li Z, Jiang L, Bai H, et al. Prevalence and clinical significance of BRAF V600E in Chinese patients with lung adenocarcinoma. Thorac Cancer 2015;6:269-74. [Crossref] [PubMed]
- Tissot C, Couraud S, Tanguy R, et al. Clinical characteristics and outcome of patients with lung cancer harboring BRAF mutations. Lung Cancer 2016;91:23-8. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical Features and Outcome of Patients With Non-Small-Cell Lung Cancer Harboring BRAF Mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Rasmussen SA, Friedman JM. NF1 gene and neurofibromatosis 1. Am J Epidemiol 2000;151:33-40. [Crossref] [PubMed]
- Cichowski K, Shih TS, Schmitt E, et al. Mouse models of tumor development in neurofibromatosis type 1. Science 1999;286:2172-6. [Crossref] [PubMed]
- Redig AJ, Capelletti M, Dahlberg SE, et al. Clinical and Molecular Characteristics of NF1-Mutant Lung Cancer. Clin Cancer Res 2016;22:3148-56. [Crossref] [PubMed]
- Cipriani NA, Abidoye OO, Vokes E, et al. MET as a target for treatment of chest tumors. Lung Cancer 2009;63:169-79. [Crossref] [PubMed]
- Ma PC, Maulik G, Christensen J, et al. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev 2003;22:309-25. [Crossref] [PubMed]
- Landi L, Minuti G, D’Incecco A, et al. Targeting c-MET in the battle against advanced nonsmall-cell lung cancer. Curr Opin Oncol 2013;25:130-6. [Crossref] [PubMed]
- Robinson KW, Sandler AB. The Role of MET Receptor Tyrosine Kinase in Non-Small Cell Lung Cancer and Clinical Development of Targeted Anti-MET Agents. Oncologist 2013;18:115-22. [Crossref] [PubMed]
- Álvarez FV, Trueba IM, Sanchis JB, et al. Recommendations of the Spanish Society of Pneumology and Thoracic Surgery on the diagnosis and treatment of non-small-cell lung cancer. Arch Bronconeumol 2016;52:2-62. [PubMed]
- Korpanty GJ, Graham DM, Vincent MD, et al. Biomarkers That Currently Affect Clinical Practice in Lung Cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol 2014;4:204. [Crossref] [PubMed]
- Kerr KM. Precision medicine in NSCLC and pathology : how does ALK fit in the pathway ? Ann Oncol 2016;27:iii16-iii24. [Crossref] [PubMed]
- Le T, Gerber DE. ALK alterations and inhibition in lung cancer. Semin Cancer Biol 2017;42:81-88. [Crossref] [PubMed]
- Tao H, Cai Y, Shi L, et al. Analysis of clinical characteristics and prognosis of patients with anaplastic lymphoma kinase-positive and surgically resected lung adenocarcinoma. Thorac Cancer 2017;8:8-15. [Crossref] [PubMed]
- Incharoen P, Reungwetwattana T, Saowapa S, et al. ALK-rearranged pulmonary adenocarcinoma in Thai Patients: From diagnosis to treatment efficacy. World J Surg Oncol 2016;14:139. [Crossref] [PubMed]
- Camidge DR, Kono SA, Flacco A, et al. Optimizing the Detection of Lung Cancer Patients Harboring Anaplastic Lymphoma Kinase (ALK) Gene Rearrangements Potentially Suitable for ALK Inhibitor Treatment. Clin Cancer Res 2010;16:5581-90. [Crossref] [PubMed]
- Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015;4:156-64. [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1 -Rearranged Non-Small-Cell Lung Cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Davies KD, Doebele RC. Molecular Pathways: ROS1 Fusion Proteins in Cancer. Clin Cancer Res 2013;19:4040-5. [Crossref] [PubMed]
- Villar Álvarez F, Muguruza Trueba I, Belda Sanchis J, et al. Executive summary of the SEPAR recommendations for the diagnosis and treatment of non-small cell lung cancer. Arch Bronconeumol 2016;52:378-88. [Crossref] [PubMed]
- Lazarus DR, Ost DE. How and when to use genetic markers for nonsmall cell lung cancer. Curr Opin Pulm Med 2013;19:331-9. [PubMed]
- Planchard D, Besse B, Groen HJ, et al. Dabrafenib plus trametinib in patients with previously treated BRAFV600E-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. [Crossref] [PubMed]
- Monsó E, Montuenga LM, Sánchez de Cos J, et al. Biological Marker Analysis as Part of the CIBERES-RTIC Cancer-SEPAR Strategic Project on Lung Cancer. Arch Bronconeumol 2015;51:462-7. [Crossref] [PubMed]
- Fernandez-Bussy S, Labarca G, Pires Y, et al. Análisis moleculares de EGFR, mutación de resistencia al EGFR, ALK y ROS1 en muestras obtenidas mediante PATB-USEB en Chile. Arch Bronconeumol 2017;53:172-4. [PubMed]
- Jeyabalan A, Bhatt N, Plummeridge M, et al. Adequacy of endobronchial ultrasound-guided transbronchial needle aspiration samples processed as histopathological samples for genetic mutation analysis in lung adenocarcinoma. Mol Clin Oncol 2016;4:119-25. [PubMed]
- Trisolini R, Cancellieri A, Tinelli C, et al. Randomized Trial of Endobronchial Ultrasound-Guided Transbronchial Needle Aspiration With and Without Rapid On-site Evaluation for Lung Cancer Genotyping. Chest 2015;148:1430-7. [Crossref] [PubMed]
- Bravaccini S, Tumedei MM, Ulivi P, et al. ALK translocation detection in non-small cell lung cancer cytological samples obtained by TBNA or EBUS-TBNA. Cytopathology 2016;27:103-7. [Crossref] [PubMed]
- Chalela R, Sánchez-Font A, Domínguez-Álvarez M, et al. Role of endobronchial ultrasound-guided transbronchial needle aspiration in the diagnosis of mediastinal tuberculosis. Med Clin (Barc) 2016;146:532-5. [Crossref] [PubMed]
- Hopkins E, Moffat D, Parkinson I, et al. Cell block samples from endobronchial ultrasound transbronchial needle aspiration provide sufficient material for ancillary testing in lung cancer -a quaternary referral centre experience. J Thorac Dis 2016;8:2544-50. [Crossref] [PubMed]
- Reynolds JP, Tubbs RR, Minca EC, et al. EGFR mutational genotyping of liquid based cytology samples obtained via fine needle aspiration (FNA) at endobronchial ultrasound of non-small cell lung cancer (NSCLC). Lung Cancer 2014;86:158-63. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Wu YL, Zhou C, Liam CK, et al. First-line erlotinib versus gemcitabine/cisplatin in patients with advanced EGFR mutation-positive non-small-cell lung cancer: analyses from the phase III, randomized, open-label, ENSURE study. Ann Oncol 2015;26:1883-9. [Crossref] [PubMed]
- Yang JC, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Mayor S. Osimertinib effective in EGFR T790M-positive lung cancer. Lancet Oncol 2017;18:e9. [Crossref] [PubMed]
- Liao BC, Lin CC, Lee JH, et al. Update on recent preclinical and clinical studies of T790M mutant-specific irreversible epidermal growth factor receptor tyrosine kinase inhibitors. J Biomed Sci 2016;23:86. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus Chemotherapy in Advanced ALK -Positive Lung Cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Chuang JC, Neal JW. Crizotinib as first line therapy for advanced ALK-positive non-small cell lung cancers. Transl lung cancer Res 2015;4:639-41. [PubMed]
- Shaw AT, Engelman JA. Ceritinib in ALK -Rearranged Non-Small-Cell Lung Cancer. N Engl J Med 2014;370:2537-9. [Crossref] [PubMed]
- Gadgeel SM, Gandhi L, Riely GJ, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol 2014;15:1119-28. [Crossref] [PubMed]
- Shaw AT, Gandhi L, Gadgeel S, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol 2016;17:234-42. [Crossref] [PubMed]
- Drilon A, Cappuzzo F, Ou SH, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15-26. [Crossref] [PubMed]
- Pardoll D. Cancer Immunotherapy with Vaccines and Checkpoint Blockade. In: Mendelsohn J, Howley PM, Israel MA, et al. editors.The Molecular Basis of Cancer. Fourth Edition. Atlanta: Elsevier Inc., 2014:709-38.
- Naylor EC, Desani JK, Chung PK. Targeted Therapy and Immunotherapy for Lung Cancer. Surg Oncol Clin N Am 2016;25:601-9. [Crossref] [PubMed]
- Du L, Herbst RS, Morgensztern D. Immunotherapy in Lung Cancer. Hematol Oncol Clin North Am. 2017;31:131-41. [Crossref] [PubMed]
- Vansteenkiste J, Zielinski M, Linder A, et al. Adjuvant MAGE-A3 immunotherapy in resected non-small-cell lung cancer: phase II randomized study results. J Clin Oncol 2013;31:2396-403. [Crossref] [PubMed]
- Butts C, Socinski MA, Mitchell PL, et al. Tecemotide (L-BLP25) versus placebo after chemoradiotherapy for stage III non-small-cell lung cancer (START): a randomised, double-blind, phase 3 trial. Lancet Oncol 2014;15:59-68. [Crossref] [PubMed]
- Nemunaitis J, Nemunaitis M, Senzer N, et al. Phase II trial of Belagenpumatucel-L, a TGF-beta2 antisense gene modified allogeneic tumor vaccine in advanced non small cell lung cancer (NSCLC) patients. Cancer Gene Ther 2009;16:620-4. [Crossref] [PubMed]
- Khanna P, Blais N, Gaudreau PO, et al. Immunotherapy Comes of Age in Lung Cancer. Clin Lung Cancer 2017;18:13-22. [Crossref] [PubMed]
- Zatloukal P, Heo DS, Park K, et al. Randomized phase II clinical trial comparing tremelimumab (CP-675,206) with best supportive care (BSC) following first-line platinum-based therapy in patients (pts) with advanced non-small cell lung cancer (NSCLC). 2009 ASCO Annual Meeting 2009:abstract 8071.
- Planchard D, Yokoi T, McCleod MJ, et al. A Phase III Study of Durvalumab (MEDI4736) With or Without Tremelimumab for Previously Treated Patients With Advanced NSCLC: Rationale and Protocol Design of the ARCTIC Study. Clin Lung Cancer 2016;17:232-236.e1. [Crossref] [PubMed]
- Antonia SJ, Goldberg SB, Balmanoukian AS, et al. Phase Ib study of MEDI4736, a programmed cell death ligand-1 (PD-L1) antibody, in combination with tremelimumab, a cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) antibody, in patients with advanced NSCLC. J Clin Oncol 2015.33. abstract 3014.
- Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480-9. [Crossref] [PubMed]
- Gettinger SN, Horn L, Gandhi L, et al. Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2015;33:2004-12. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Langer CJ, Gadgeel SM, Borghaei H, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol 2016;17:1497-508. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]