Genetic susceptibility to lung cancer and co-morbidities
Introduction
Lung cancer is a leading cause of cancer death and disease burden in many countries (1,2). Despite the great progress made in several areas of oncology, the treatment and outcome of lung cancer has not improved significantly. Understanding of the biological pathways involved in lung cancer aetiology is required to identify key biomolecules that could be of significant clinical value, either as predictive, prognostic or diagnostic markers, or as targets for the development of novel therapies to treat this disease, in addition to smoking avoidance strategies.
Lung cancers, like all human tumours, are caused by abnormalities in DNA sequence or expression. Lung cancer is comprised of two main histologic subtypes: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC can now be classified by ‘driver’ mutations that occur in multiple oncogenes (3-5). However, much interest remains in the genetic susceptibility to lung cancer related to single nucleotide polymorphisms (SNPs) in the germline (6,7), independent of somatic mutations that develop in the tumour.
This review will explore the latest concepts of genetic susceptibility to lung cancer. First, principles of study designs for genetic susceptibility will be outlined, particularly genome-wide association studies (GWAS), which have provide significant progress in the past 5 or more years in the field of lung cancer genetics. Next, GWAS reported for lung cancer will be described, together with candidate susceptibility genes discovered to date. Finally, GWAS for susceptibility to cigarette smoking behaviour and chronic obstructive pulmonary disease (COPD) will be summarised, as related genetic markers for the most common cause of lung cancer (cigarette smoking) and a chronic disease risk factor for lung cancer (COPD).
Genetic susceptibility to lung cancer–study designs
The heritability of lung cancer susceptibility has been clearly established in numerous studies, including analyses of familial risk (8) and segregation analyses (9). However genetic influence on lung cancer is moderate at best. For example, using the 9.6 million subject Swedish Family-Cancer database, Czene et al. estimated heritability at 8% (10) and in twin studies and a higher concordance for monozygotic than for dizygotic twins has been noted (11). With tobacco smoking being by far the strongest environmental cause, it is possible that the heritable effects of genes governing smoking behaviour, [given the high heritability of smoking habits, :0.5 in twin studies (12)], rather those determining individual susceptibility to carcinogenesis may play a more important role. However Lorenzo Bermejo et al. estimated the relative risk of lung cancer attributable to smoking according to the extent to which smokers transmit their smoking habits to the offspring (heritability of smoking), the prevalence of smoking in the general population, and the risk of lung cancer for smokers compared with non-smokers (13). They showed that the relative risk of lung cancer for the offspring of lung cancer patients attributable to smoking was 1.19 when published data on smoking practice were modelled suggesting familial cases of lung cancer cannot be attributed to shared smoking habits.
Given that there are apparent genetic determinants of lung cancer, there are a number of alternative study approaches that can be utilised to determine the genetic determinants of disease susceptibility.
Linkage analysis involves proposing a model to explain the inheritance pattern of phenotypes and genotypes observed in a pedigree (14). Linkage is evident when a gene that produces a phenotypic trait and its surrounding markers are coinherited. In contrast, those markers not associated with the anomalous phenotype of interest will be randomly distributed among affected family members as a result of the independent assortment of chromosomes and crossing over during meiosis. Association studies do not examine inheritance patterns of alleles; rather, they are case-control studies based on a comparison of allele frequencies between groups of affected and unaffected individuals from a population. A particular allele is said to be associated with the trait if it occurs at a significantly higher frequency among affected individuals as compared with those in the control group. The odds ratio of the trait in individuals is then assessed as the ratio of the frequency of the allele in the affected population compared with the unaffected population. The greatest problem in association studies is the selection of a suitable control group to compare with the affected population group. Genome-wide linkage analysis in family cohorts resulted in the identification of highly penetrant, low-frequency susceptibility genes for cancers, such as BRCA1 and BRCA2 for breast cancer and APC for colorectal cancer.
For lung cancer, several studies have attempted to identify susceptibility loci using a genome-wide linkage approach. However, while a few genetic loci that potentially harbour susceptibility genes have been identified, e.g., linkage of lung, laryngeal, and pharyngeal cancer in families to a region on chromosome 6q23-25 (15), no causal gene has been identified and, as with subsequent GWAS (see below), there is considerable overlap between the result for lung cancer and those for COPD, and lung function (16).
GWAS
Advances in array-based SNP genotyping technologies and haplotype mapping of the human genome (17) have presented the possibility of simultaneously determining millions of SNPs throughout the genome of an individual and this has allowed extension of association study design to allow hypothesis independent assessment of association across the genome. GWAS have revolutionized the study of genetic factors in complex common disease (18,19). For more than 200 phenotypes, from common diseases to physiological measurements such as height and BMI and biological measurements such as circulating lipid levels, GWAS have provided compelling statistical associations for over hundreds of different loci in the human genome (www.gwascentral.org/). There are now clearly established approaches for GWAS including stringent genotype calling, quality control, population stratification (genomic controls) and statistical techniques (20). Due to the large number of statistical tests undertaken, carefully controlling for multiple testing using Bonferroni or false discovery rate (FDR) corrections is essential. A cluster of P-values below the 1% FDR from SNPs in one chromosomal location is defined as the region of ‘maximal association’ and is the first candidate gene region to examine further with analysis of secondary outcome measures, gene database searches, fine mapping to find the causal locus and replication in other cohorts/populations. It is unlikely that the SNP showing the strongest association will be the causal locus, as SNPs are chosen to provide maximal coverage of other variation in that region of the genome and not on biological function. Once a candidate region or gene is identified, gene expression can be compared between a selection of cases and controls and within individuals of different genotypes to provide further evidence for the genes involvement in disease. If linkage disequilibrium prevents the identification of a specific gene in a haplotype block then it may be necessary to utilize different racial and ethnic populations to hone in on the causative candidate gene that accounts for the genetic signal in GWAS (21).
Valuable insights into lung cancer susceptibility genes have been identified using the GWAS approach; however, the loci identified account for an extremely small proportion of the familial risk. The finding that loci identified through GWAS studies for common disease fail to account for all heritability of these conditions (termed missing heritability) is a subject of much discussion. There are a number of possible reasons that may account for this observation. These include gene-gene interaction, gene-environment interaction, and other types of genetic variation such as rare variants and structural variation and epigenetic heritability. In the future, the analysis of genome-wide copy number variation and/or rare variants through exome- or whole-genome sequencing, as is being applied to other complex diseases, may identify further loci responsible for inherited susceptibility to lung cancer (22). However one approach is to utilise novel analytical approaches to identify weakly associated variants whose effect may be lost in the GWAS approach due to the stringent significance level after multiple comparison correction. For example, utilisation of a less stringent multiple correction followed by gene pathway analysis can highlight genes involved in common biological pathways in the ‘grey area’ of SNPs whose association with disease status lies below the conventional level of genome-wide significance. Using a similar approach Zhang et al. performed a two-stage pathway analysis in GWAS of lung cancer in Han Chinese using gene set enrichment analysis (GSEA) method. Four pathways (achPathway, metPathway, At1rPathway and rac1Pathway) were consistently significant and may provide new insights into the etiology of lung cancer (23).
GWAS of lung cancer
GWAS of susceptibility to lung cancer in smokers
A number of GWAS have now been performed in a range of populations, to test genetic influences in susceptibility to lung cancer (Table 1).
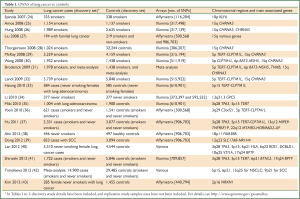
Full Table
An initial, relatively small GWAS was reported in an Italian population, showing association with SNPs in the KLF6 gene, but not in replication cohorts (24). Another relatively small study of patients with familial lung cancer found associations with SNPs at chromosomal region 15q (27). Larger scale GWAS have since been performed in Caucasian populations, with replication cohorts (25,26,28-30,32) and meta-analyses (31,32,42). These GWAS have found statistically significant associations with SNPs, particularly in chromosomal regions 15q, 5p and 6p (Table 1). GWAS have also been undertaken in Asian populations with lung cancer (35-37,39,41), identifying some similar SNPs as detected in the studies of Caucasian populations, but also finding other SNPs conferring lung cancer risk distinctly in Asian populations (44).
In many of these studies, the observed associations with key SNPs were independent of smoking status or smoking history (25,26,29,32), although in some studies, SNPs (e.g., on 15q) were related to smoking behaviour (28). In studies of smokers, population attributable risk (PAR) for lung cancer for these SNPs, where calculated, were modest, ranging between 14% (26) and 18% (28), compared to the overwhelming PAR of >80% from tobacco smoking.
GWAS in specific lung cancer populations
GWAS have also been undertaken in never smokers, and also to address other lung cancer-related questions. A GWAS of lung cancer in never smokers in the USA, with replication cohorts, identified an association with SNPs at the 13q region, in the GPC5 gene (34). Studies have also examined genome-wide association in never smokers in Asian populations (38,40,43), finding new susceptibility loci that were different to the loci found for Caucasian populations. SNPs in the TERT gene on 5p15 were associated with lung adenocarcinoma as a specific lung cancer histology (32), whereas SNPs at 9p21 are associated with risk for lung squamous cell carcinoma (SCC) (42). Genome-environment interaction was tested in a GWAS of lung cancer risk and self-reported asbestos exposure (45). Whilst this pilot study was not sufficiently powered to find significant differences, a suggested gene-asbestos exposure interaction was seen for SNPs in C7orf54 on 7q32. In addition, a number of GWAS have identified SNPs that predict response to chemotherapy in patients with SCLC (46) and NSCLC (47), and other GWAS have explored SNPs related to prognosis and survival in patients with lung cancer [e.g., (48)].
Candidate genes for lung cancer susceptibility from GWAS
The discovery and replication studies from the GWAS (Table 1), and other replication studies since, have identified emerging patterns in candidate genes for lung cancer susceptibility [reviewed in (6,7,42)]. Consistent candidate genes for Caucasian smoking populations have been the neuronal nicotinic acetylcholine receptor (nAChR) subunits (cholinergic receptor, nicotinic, alpha 3 and 5: CHRNA3 and CHRNA5) at 15q25. Neuronal nAChRs are activated by acetylcholine or nicotine, and comprise subunits (pentamers) of α and β subunits. In the lungs, nAChRs are expressed in neurones, and also non-neuronal cells, including bronchial epithelial cells and lung cancer cells. Whilst SNPs in nAChRs may alter risk of lung cancer through smoking behaviour, these SNPs could also regulate direct effects of nicotine, through anti-apoptotic and proliferative effects, or effects of nicotine-derived carcinogens in tobacco smoke (6,7,49-51).
On the 5p15 locus, SNPs in the TERT and CLPTM1L genes have been associated in a number of GWAS of lung cancer and other cancers (52). The TERT gene encodes human telomerase reverse transcriptase, which is important in the maintenance of telomere length. CLPTM1L (cleft lip and palate transmembrane protein 1-like protein) may induce apoptosis in lung cells (29). The identified SNPs in TERT have generally been intronic, and in linkage disequilibrium with SNPs in CLPTM1L. On 6p21, BAT3 and MSH5 have emerged as signals in a number of GWAS. BAT3 (renamed BAG6, BCL2-associated athanogene 6) encodes a nuclear protein involved in DNA damage-induced apoptosis and modulation of p53 in response to genotoxic stress (7). The MSH5 [mutS homolog 5 (E. coli)] gene is involved in DNA mismatch repair.
Whilst the 5p15 SNPs have demonstrated replication in both Caucasian and Asian populations, this has not been the case for the 15q SNPs (40,53). This discordance may represent differences in lung cancer aetiology between smoking and never smoking populations (particularly where indoor air pollution from biomass fuels may be the predominant carcinogen). In the never smokers, especially in Asian female populations, other candidate genes emerge, for example, that are involved in receptor tyrosine kinase activity [ROS1, c-ros oncogene 1, receptor tyrosine kinase (40)], vesicle transport [VTI1A, vesicle transport through interaction with t-SNAREs 1A (40)] and cell adhesion [NRXN1, neurexin 1 (43)]. Unexpectedly, the transcription factor TP63 (tumor protein p63) is also a candidate marker in the never smoking populations. TP63 encodes a protein which is often used as an immunohistochemical marker of squamous cell carcinoma, a cancer strongly associated with tobacco smoking (54,55).
Many of these associations are novel for lung cancer susceptibility, and were not detected in previous candidate gene studies of lung cancer, which focused on metabolising enzymes [e.g., CYP1A1 (56-58)], oxidative stress pathways and other DNA repair mechanisms (7).
GWAS and lung cancer pharmacogenetics
The GWAS approach is now being extended to examine other related phenotypes in lung cancer. For example, Han et al. recently undertook a GWAS of survival in small-cell lung cancer patients treated with irinotecan plus cisplatin chemotherapy, and identified candidate SNPs that may be predictive of the clinical outcome (59).
GWAS of smoking behaviour
Of relevance to lung cancer aetiology, GWAS of smoking behaviour have been performed in large population cohorts (Table 2), and have focused on smoking initiation, smoking quantity (cigarettes per day) and success of smoking cessation. The interest for lung cancer susceptibility is not only for causation from smoking, but also for SNPs that are common to both smoking behaviour and lung cancer, that could have direct biological effects.

Full Table
Smoking onset
Onset of smoking has been associated with the 11p region, with BDNF (brain-derived neurotrophic factor), a neurotropin, identified as a possible candidate gene (61). Other regions identified with age of smoking initiation in a study of patients with COPD were 6p21 (HLA) and 2q21 (intergenic region) (63).
Smoking quantity
The intensity of smoking (smoking quantity) has been consistently associated in GWAS with SNPs in the 15q25 region (60-65), at the loci of nAChR genes (especially CHRNA3 and CHRNA5). Other loci include 19q (CYP2A6, a cytochrome P450 nicotine metabolising enzyme) (61-63, 66) and 8p (CHRNB3, a nAChR subunit) (62).
Smoking cessation
A range of biopsychosocial factors influence an individual’s ability to abstain from smoking (67). Genetic factors for smoking cessation identified from GWAS have centred around SNPs in the 9q region, including DBH (dopamine beta-hydroxylase) (61,63), which is involved in the metabolism of dopamine.
GWAS of COPD
Lung cancer and COPD frequent coexist in at-risk smokers. Epidemiological evidence supports an association between the presence of COPD and increased risk of developing lung cancer. Common mechanisms for susceptibility to lung cancer and COPD, in addition to tobacco smoke, may involve biological processes such as inflammation, epithelial-mesenchymal transition, abnormal repair, oxidative stress, and cell proliferation. In addition, genomic and epigenomic changes are likely to alter biological pathways, leading to susceptibility to lung cancer and COPD (68,69). Therefore, studying genetic influences in COPD could yield greater insight into the shared pathogenesis of lung cancer and COPD. Importantly, genetic epidemiological principles should be considered when designing and interpreting studies of COPD and lung cancer, because of the shared aetiological and possibly genetic factors (70).
GWAS of susceptibility to COPD
A number of GWAS have been performed for COPD (Table 3), albeit a smaller number of studies than the lung cancer GWAS to date.
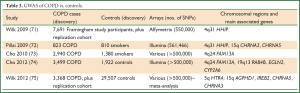
Full Table
GWAS have so far been undertaken in the Framingham cohort (71); Bergen (Norway) COPD Cohort (with replication in other cohorts) (72); combined Bergen cohort with National Emphysema Treatment Trial (NETT) and Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) study subjects (73); combined four cohorts [ECLIPSE, Normative Aging Study, Bergen (Norway) COPD Cohort and COPD Gene] (74); and a combination of 15 cohorts (75).
Candidate genes for COPD from GWAS
Several regions significantly associated with COPD have emerged from the COPD GWAS to date (Table 3). At the 15q locus (similar to the GWAS for lung cancer and for smoking behaviour), SNPs in the nAChR subunit genes (CHRNA5, CHRNA3) were associated with COPD (72,75), possibly indicating a link with smoking intensity as an aetiological factor, although direct effects should also be considered (76). Similarly, association at 19q13 may be related to smoking behaviour [e.g., CYP2A6 (74)].
Other novel associations have been found for COPD in these GWAS. At 4q31, hedgehog interacting protein (HHIP) has been identified as a candidate in two studies (71,72). HHIP encodes a membrane glycoprotein that is an inhibitor of hedgehog signalling, which is involved in development processes. Gene expression studies in BEAS-2B bronchial epithelial cell lines implicate HHIP in extracellular matrix and cell proliferation (77). At 4q24, FAM13A (family with sequence similarity 13, member A) has been detected in two GWAS (73,74), and also in a genetic association in which SNPs were related to both COPD and lung cancer, indicating a possible shared genetic pathway (78). FAM13A contains a Rho GTPase-activating protein-binding domain, inhibits signal transduction and responds to hypoxia; however, its full function in the lung remains to be determined. At the 5q region, HTR4 [5-hydroxytryptamine (serotonin) receptor 4] was associated with COPD in smokers; its function in airways disease may involve regulation of cytokine release (75).
Summary and clinical implications
Main findings to date from GWAS
Whilst lung cancer is predominantly caused by cigarette smoking, a genetic component to susceptibility is well-recognised in epidemiological studies. GWAS have now been completed in lung cancer and the related phenotypes of smoking behaviour and COPD. Large scale, multi-cohort GWAS of lung cancer in mainly Caucasian, smoking populations have identified strong associations for lung cancer mapped to chromosomal regions 15q (CHRNA3, CHRNA5), 5p (TERT-CLPTM1L locus) and 6p (BAT3-MSH5). Some studies in Asian populations of smokers have found similar risk loci, whereas other GWAS, particularly in never smoking Asian females, have identified associations in other chromosomal regions that are distinct from the smoking-related genetic loci. GWAS of smoking behaviour have identified risk loci for smoking quantity at 15q (similar genes to lung cancer susceptibility: CHRNA3, CHRNA5) and also at 19q (CYP2A6). Other genes have been mapped for smoking initiation and smoking cessation. In COPD, GWAS in large cohorts have also found NAChR SNPs mapping at 15q as risk loci, as well as other regions at 4q31 (HHIP), 4q24 (FAM13A) and 5q (HTR4). The overlap in risk loci between lung cancer, smoking behaviour and COPD may be due to the effects of nicotine addiction; However, more work needs to be undertaken to explore the potential direct effects of nicotine and its metabolites in gene-environment interaction in these phenotypes.
Applications and future directions
From the evidence presented to date, GWAS have been useful not only in addressing genetic influences in lung cancer susceptibility, but also gene-environment interaction in terms of smoking as causation, as well as COPD as a risk factor for lung cancer. The translation of findings from the lung cancer and related GWAS could in the future enable profiling of an individual’s risk of lung cancer, biomarkers for diagnosis, and markers for prognosis and therapy. The challenge now will be to combine genomics (79), epigenomics (80), transcriptomics (81-83) and proteomics profiling to improve the management of patients with lung cancer and related comorbidities (68). Future studies should include DNA sequencing of lung tumours and lung tissue, and network analysis of genomic information and clinical phenotypes (3,84,85). Goals of future genetic susceptibility studies of lung cancer should now be focused on refining the strongest risk loci in a wide range of populations with lung cancer, and integrating other clinical and biomarker information, in order to achieve the aim of personalised therapy for lung cancer (4), through enhanced diagnosis, prognosis, prevention and management.
Acknowledgements
This work was supported by Asthma, Allergy & Inflammation Research (AAIR) (JWH), NHMRC Career Development Fellowship (IY), NHMRC Practitioner Fellowship (KF), and project grants from NHMRC, Queensland Health and Medical Research, The Prince Charles Hospital Foundation, and Cancer Council Queensland.
Disclosure: The authors declare no conflict of interest.
References
- Thun MJ, Lally CA, Flannery JT, et al. Cigarette smoking and changes in the histopathology of lung cancer. J Natl Cancer Inst 1997;89:1580-6. [PubMed]
- Mathers C, Vos T. The burden of disease and injury in australia-summary report. Canberra: Australian Institute of Health and Welfare, 1999.
- Sriram KB, Larsen JE, Yang IA, et al. Genomic medicine in non-small cell lung cancer: paving the path to personalized care. Respirology 2011;16:257-63. [PubMed]
- Fong KM, Yang IA, Bowman RV. Personalized medicine for lung cancer. Lung Cancer Management 2012;1:83-6.
- Sriram KB, Tan ME, Savarimuthu SM, et al. Screening for activating EGFR mutations in surgically resected nonsmall cell lung cancer. Eur Respir J 2011;38:903-10. [PubMed]
- Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol 2011;12:399-408. [PubMed]
- Marshall AL, Christiani DC. Genetic susceptibility to lung cancer--light at the end of the tunnel? Carcinogenesis 2013;34:487-502. [PubMed]
- Jonsson S, Thorsteinsdottir U, Gudbjartsson DF, et al. Familial risk of lung carcinoma in the Icelandic population. JAMA 2004;292:2977-83. [PubMed]
- Sellers TA, Chen PL, Potter JD, et al. Segregation analysis of smoking-associated malignancies: evidence for Mendelian inheritance. Am J Med Genet 1994;52:308-14. [PubMed]
- Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer 2002;99:260-6. [PubMed]
- Braun MM, Caporaso NE, Page WF, et al. A cohort study of twins and cancer. Cancer Epidemiol Biomarkers Prev 1995;4:469-73. [PubMed]
- Tyndale RF. Genetics of alcohol and tobacco use in humans. Ann Med 2003;35:94-121. [PubMed]
- Lorenzo Bermejo J, Hemminki K. Familial lung cancer and aggregation of smoking habits: a simulation of the effect of shared environmental factors on the familial risk of cancer. Cancer Epidemiol Biomarkers Prev 2005;14:1738-40. [PubMed]
- Peddle L, Rahman P. Genetic epidemiology of complex phenotypes. Methods Mol Biol 2009;473:187-201. [PubMed]
- Bailey-Wilson JE, Amos CI, Pinney SM, et al. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet 2004;75:460-74. [PubMed]
- Schwartz AG, Ruckdeschel JC. Familial lung cancer: genetic susceptibility and relationship to chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2006;173:16-22. [PubMed]
- International HapMap Consortium. A haplotype map of the human genome. Nature 2005;437:1299-320. [PubMed]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science 2008;322:881-8. [PubMed]
- McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356-69. [PubMed]
- Amos CI. Successful design and conduct of genome-wide association studies. Hum Mol Genet 2007;16 Spec No. 2:R220-5.
- Lam AC, Powell J, Wei WH, et al. A combined strategy for quantitative trait loci detection by genome-wide association. BMC Proc 2009;3 Suppl 1:S6.
- Frazer KA, Murray SS, Schork NJ, et al. Human genetic variation and its contribution to complex traits. Nat Rev Genet 2009;10:241-51. [PubMed]
- Zhang R, Zhao Y, Chu M, et al. Pathway analysis for genome-wide association study of lung cancer in Han Chinese population. PLoS One 2013;8:e57763. [PubMed]
- Spinola M, Leoni VP, Galvan A, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett 2007;251:311-6. [PubMed]
- Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40:616-22. [PubMed]
- Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452:633-7. [PubMed]
- Liu P, Vikis HG, Wang D, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst 2008;100:1326-30. [PubMed]
- Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638-42. [PubMed]
- McKay JD, Hung RJ, Gaborieau V, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet 2008;40:1404-6. [PubMed]
- Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 2008;40:1407-9. [PubMed]
- Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res 2009;69:6633-41. [PubMed]
- Landi MT, Chatterjee N, Yu K, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009;85:679-91. [PubMed]
- Hsiung CA, Lan Q, Hong YC, et al. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet 2010;6:e1001051. [PubMed]
- Li Y, Sheu CC, Ye Y, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol 2010;11:321-30. [PubMed]
- Miki D, Kubo M, Takahashi A, et al. Variation in Tp63 Is Associated with Lung Adenocarcinoma Susceptibility in Japanese and Korean Populations. Nat Genet 2010;42:893-6. [PubMed]
- Yoon KA, Park JH, Han J, et al. A genome-wide association study reveals susceptibility variants for non-small cell lung cancer in the Korean population. Hum Mol Genet 2010;19:4948-54. [PubMed]
- Hu Z, Wu C, Shi Y, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011;43:792-6. [PubMed]
- Ahn MJ, Won HH, Lee J, et al. The 18p11.22 locus is associated with never smoker non-small cell lung cancer susceptibility in Korean populations. Hum Genet 2012;131:365-72. [PubMed]
- Dong J, Jin G, Wu C, et al. Genome-wide association study identifies a novel susceptibility locus at 12q23.1 for lung squamous cell carcinoma in han chinese. PLoS Genet 2013;9:e1003190. [PubMed]
- Lan Q, Hsiung CA, Matsuo K, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet 2012;44:1330-5. [PubMed]
- Shiraishi K, Kunitoh H, Daigo Y, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet 2012;44:900-3. [PubMed]
- Timofeeva MN, Hung RJ, Rafnar T, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet 2012;21:4980-95. [PubMed]
- Kim JH, Park K, Yim SH, et al. Genome-wide association study of lung cancer in korean non-smoking women. J Korean Med Sci 2013;28:840-7. [PubMed]
- Dong J, Hu Z, Wu C, et al. Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat Genet 2012;44:895-9. [PubMed]
- Wei S, Wang LE, McHugh MK, et al. Genome-wide gene-environment interaction analysis for asbestos exposure in lung cancer susceptibility. Carcinogenesis 2012;33:1531-7. [PubMed]
- Wu C, Xu B, Yuan P, et al. Genome-wide examination of genetic variants associated with response to platinum-based chemotherapy in patients with small-cell lung cancer. Pharmacogenet Genomics 2010;20:389-95. [PubMed]
- Wu X, Ye Y, Rosell R, et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J Natl Cancer Inst 2011;103:817-25. [PubMed]
- Lee Y, Yoon KA, Joo J, et al. Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis 2013;34:307-13. [PubMed]
- West KA, Brognard J, Clark AS, et al. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest 2003;111:81-90. [PubMed]
- Tournier JM, Birembaut P. Nicotinic acetylcholine receptors and predisposition to lung cancer. Curr Opin Oncol 2011;23:83-7. [PubMed]
- Al-Wadei HA, Al-Wadei MH, Schuller HM. Cooperative regulation of non-small cell lung carcinoma by nicotinic and beta-adrenergic receptors: a novel target for intervention. PLoS One 2012;7:e29915. [PubMed]
- Mocellin S, Verdi D, Pooley KA, et al. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst 2012;104:840-54. [PubMed]
- Truong T, Hung RJ, Amos CI, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst 2010;102:959-71. [PubMed]
- Khayyata S, Yun S, Pasha T, et al. Value of P63 and CK5/6 in distinguishing squamous cell carcinoma from adenocarcinoma in lung fine-needle aspiration specimens. Diagn Cytopathol 2009;37:178-83. [PubMed]
- Zhang P, Han Y, Huang L, et al. Expression and Clinical Significance of TTF-1 and p63 in NSCLC. Zhongguo Fei Ai Za Zhi 2009;12:995-9. [PubMed]
- Larsen JE, Colosimo ML, Yang IA, et al. Risk of non-small cell lung cancer and the cytochrome P4501A1 Ile462Val polymorphism. Cancer Causes Control 2005;16:579-85. [PubMed]
- Larsen JE, Colosimo ML, Yang IA, et al. CYP1A1 Ile462Val and MPO G-463A interact to increase risk of adenocarcinoma but not squamous cell carcinoma of the lung. Carcinogenesis 2006;27:525-32. [PubMed]
- Wright CM, Larsen JE, Colosimo ML, et al. Genetic association study of CYP1A1 polymorphisms identifies risk haplotypes in nonsmall cell lung cancer. Eur Respir J 2010;35:152-9. [PubMed]
- Han JY, Lee YS, Shin ES, et al. A genome-wide association study of survival in small-cell lung cancer patients treated with irinotecan plus cisplatin chemotherapy. Pharmacogenomics J 2013. [Epub ahead of print]. [PubMed]
- Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010;42:436-40. [PubMed]
- Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010;42:441-7. [PubMed]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010;42:448-53. [PubMed]
- Siedlinski M, Cho MH, Bakke P, et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax 2011;66:894-902. [PubMed]
- Saccone NL, Culverhouse RC, Schwantes-An TH, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet 2010;6:e1001053. [PubMed]
- David SP, Hamidovic A, Chen GK, et al. Genome-wide meta-analyses of smoking behaviors in African Americans. Transl Psychiatry 2012;2:e119. [PubMed]
- Kumasaka N, Aoki M, Okada Y, et al. Haplotypes with copy number and single nucleotide polymorphisms in CYP2A6 locus are associated with smoking quantity in a Japanese population. PLoS One 2012;7:e44507. [PubMed]
- Fung PR, Snape-Jenkinson SL, Godfrey MT, et al. Effectiveness of hospital-based smoking cessation. Chest 2005;128:216-23. [PubMed]
- Yang IA, Relan V, Wright CM, et al. Common pathogenic mechanisms and pathways in the development of COPD and lung cancer. Expert Opin Ther Targets 2011;15:439-56. [PubMed]
- Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer 2013;13:233-45. [PubMed]
- El-Zein RA, Young RP, Hopkins RJ, et al. Genetic predisposition to chronic obstructive pulmonary disease and/or lung cancer: important considerations when evaluating risk. Cancer Prev Res (Phila) 2012;5:522-7. [PubMed]
- Wilk JB, Chen TH, Gottlieb DJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 2009;5:e1000429. [PubMed]
- Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009;5:e1000421. [PubMed]
- Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet 2010;42:200-2. [PubMed]
- Cho MH, Castaldi PJ, Wan ES, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet 2012;21:947-57. [PubMed]
- Wilk JB, Shrine NR, Loehr LR, et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med 2012;186:622-32. [PubMed]
- Bossé Y. Updates on the COPD gene list. Int J Chron Obstruct Pulmon Dis 2012;7:607-31. [PubMed]
- Zhou X, Qiu W, Sathirapongsasuti JF, et al. Gene expression analysis uncovers novel hedgehog interacting protein (HHIP) effects in human bronchial epithelial cells. Genomics 2013;101:263-72. [PubMed]
- Young RP, Hopkins RJ, Hay BA, et al. FAM13A locus in COPD is independently associated with lung cancer - evidence of a molecular genetic link between COPD and lung cancer. Appl Clin Genet 2010;4:1-10. [PubMed]
- Sriram KB, Larsen JE, Savarimuthu Francis SM, et al. Array-comparative genomic hybridization reveals loss of SOCS6 is associated with poor prognosis in primary lung squamous cell carcinoma. PLoS One 2012;7:e30398. [PubMed]
- Davidson MR, Larsen JE, Yang IA, et al. Microrna-218 Is Deleted and Downregulated in Lung Squamous Cell Carcinoma. PLoS ONE 2010;5:e12560. [PubMed]
- Larsen JE, Pavey SJ, Bowman R, et al. Gene expression of lung squamous cell carcinoma reflects mode of lymph node involvement. Eur Respir J 2007;30:21-5. [PubMed]
- Larsen JE, Pavey SJ, Passmore LH, et al. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis 2007;28:760-6. [PubMed]
- Larsen JE, Pavey SJ, Passmore LH, et al. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res 2007;13:2946-54. [PubMed]
- Daniels M, Goh F, Wright CM, et al. Whole genome sequencing for lung cancer. J Thorac Dis 2012;4:155-63. [PubMed]
- Silverman EK. Perspective: how can genetics help? Nature 2012;489:S7. [PubMed]


