Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer
Introduction
Non-small cell lung cancer (NSCLC), one of histological types of lung cancer (ratio at 85:100), is divided into three subtypes, consisting of adenocarcinoma, squamous-cell carcinoma and large-cell carcinoma (1-3). Although multiple and optimal treatments, including surgery, chemotherapies and radiotherapies, have been widely utilized in NSCLC patients, 5-year overall survival (OS) rate is still low at approximate 15% (4,5), One of the possible reasons is the lack of specific biomarkers at the early stage. Thus, exploring precise biomarkers related to clinicopathological features and the prognosis for NSCLC is greatly needed to monitor the progress in NSCLC patients.
MicroRNAs (miRNAs), a great family with small non-coding RNA molecules at length about 19 to 24 nucleotides, have established their value into multiple pathological processes, including cell proliferation, cell apoptosis and cell differentiation, through the post-transcriptional regulation of genes expression (6-8). Accumulating evidences disclose that several miRNAs act as important biomarkers for the diagnosis and prognosis of various cancers, such as NSCLC, pancreatic cancer and colorectal cancer (9-11).
MiR-34 family consists of three members, including miR-34a with own coding transcript (sited on chromosome 1), and miR-34b/c with a shared primary transcript (sited on chromosome 11) (12). Dysregulated expressions of miR-34 are observed in some carcinomas, such as colorectal cancer, breast cancer or even lung cancer (13-15). Particularly, aberrant miR-34b/c expressions have been reported to involve into the prognosis of small cell lung cancer (SCLC) and it could be a potential therapeutic target in SCLC patients (16). However, little is known about the effects of miR-34 expression in NSCLC. Therefore, the purpose of this study was to evaluate the association of plasma miR-34a/b/c expressions with the clinicopathological properties and the prognosis in NSCLC patients.
Methods
Participants
A total of 196 NSCLC patients were enrolled from 2010/04/01 to 2012/03/31 at the Department of Thoracic Surgery, The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology. All patients received a complete resection and didn’t receive any preoperative adjuvant therapy as well as underwent thoracic surgery with age above 18 years, and the diagnosis of NSCLC was determined according to clinical features, radiological examination and pathological confirmation. Patients with other tumor history or malignant hematological disease history, or complicated with other lung diseases such as chronic obstructive pulmonary disease (COPD), lung infection and so on were excluded. All patients provided written informed consents and this study was approved by the Ethics Committee of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology (No. 20100213015) and was consistent with the principles outlined in the Declaration of Helsinki.
Samples
Plasma samples were collected from all patients post surgery and before any other treatments performed including chemotherapy, radiotherapy, immunotherapy and so on. Tissue samples from the tumoral area were also obtained during the surgery.
RNA extraction and real-time PCR
Total RNA was extracted from plasma and tissue samples by Trizol Reagent (Takara, Japan), and evaluated by the spectrophotometer for concentration and purity of the RNA. RNA was subsequently subjected to reverse transcription with the PrimeScript real-time reagent kit (Takara, Japan) and quantitative analysis of miR-34a/b/c expressions were detected by SYBR Premix Ex TaqTM II (Takara, Japan). All determination was carried out according to the instructions of manufacturers. Levels of miR-34/a/b/c expression were subsequently calculated utilizing the 2−ΔΔCt method and U6 was served as the internal reference.
Assessments and follow-ups
Demographic, pathological and clinical features were collected for all patients. All tumours were staged in accordance with the 7th edition of the American Joint Committee on Cancer (AJCC) cancer staging manual. The last follow-up date was 2016/08/31, and the median follow-up time was 56 months. OS and disease-free survival (DFS) were defined from the surgery to time of death from any cause and time of documented local or distant recurrence of initial cancer, respectively.
Statistics
Statistics was performed using SPSS 22.0 (IBM, USA) and 2010 office software (Microsoft, USA). Data was shown as mean value ± standard deviation, median value (25th–75th quarter), or counts (with or without percentage). Comparison was determined by Wilcoxon rank sum test or Kruskal-Wallis H rank sum test. Kaplan-Meier curves were drawn for DFS and OS, and compared by log-rank test. P≤0.05 was considered as statistically significant.
Results
Characteristics
Median value of age was 58.0 (range, 27.0–82.0) years in a total of 196 NSCLC patients, the numbers of male and female were 118 and 78, respectively (Table 1). Additionally, there were 87 (44.4%) patients with adenocarcinoma, 96 (49.0%) patients with squamous cell carcinoma, and others were 13 (6.6%). According to pathological grade, the numbers of NSCLC patients with poor, moderate and well differentiation were 52 (26.5%), 97 (49.5%) and 47 (24.0%), respectively. In plasma sample, the median values of miR-34a/b/c expressions were observed to be 0.632 (0.428–0.866), 0.786 (0.480–1.008) and 0.342 (0.270–0.426), while the median levels of tumor tissue miR-34a/b/c expressions were 1.323 (0.750–2.204), 1.092 (0.537–1.849) and 0.682 (0.304–0.949), respectively. Other clinical and pathological characteristics of NSCLC patients were presented in Table 1.
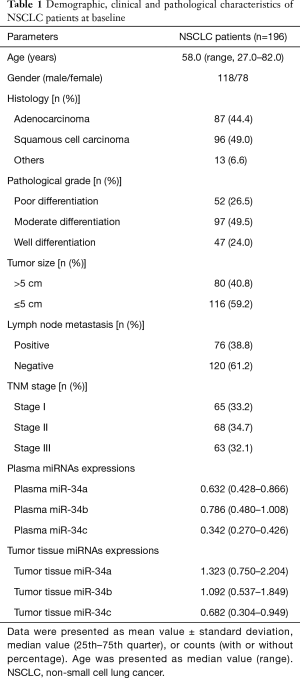
Full table
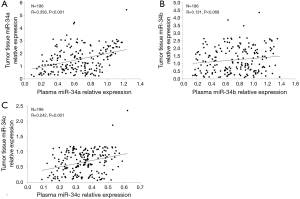
Correlation between miR-34a/b/c expressions in plasma and in tumor tissues
Correlation between miR-34a/b/c expressions in plasma and tumor tissues was analyzed in Figure 1. MiR-34a and miR-34c levels in plasma were positively associated with that in tumor tissues (P<0.001 and P=0.001, respectively). However, no correlation of miR-34b in plasma with that in tumor tissues was found in this study (P=0.068). These indicated that miR-34a/c expressions have great consistency between plasma and tumor tissues.
Comparison of plasma miR-34a/b/c expressions between subgroups divided by clinicopathological features
Wilcoxon rank sum test or Kruskal-Wallis H rank sum test were used to compare plasma miR-34a/b/c expressions between subgroups divided by clinicopathological features (Table 2). Plasma miR-34a expression was negatively correlated with lymph node metastasis (P=0.002). Possible correlations of plasma miR-34a with TNM stage (P=0.068), plasma miR-34b with lymph node metastasis (P=0.054) and plasma miR-34c with histology (P=0.064) were found but without statistical significance. No difference of plasma miR-34a/b/c expressions between subgroups divided by other clinicopathological features was observed in Table 2.
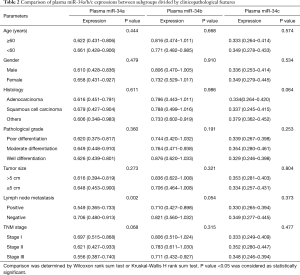
Full table
Comparison of tumor tissue miR-34a/b/c expressions between subgroups divided by clinicopathological features
In terms of tumor tissue sample, miR-34a expression was negatively associated with lymph node metastasis (P=0.018) (Table 3). MiR-34a expression seemed to be negatively correlated with TNM stage (P=0.096), and miR-34c expression was likely to be associated with age (≥60 years) (P=0.092), gender (female) (P=0.064), negative lymph node metastasis (P=0.097) but without statistical significance. No difference of plasma miR-34a/b/c expressions between subgroups divided by other clinicopathological features was presented in Table 3.
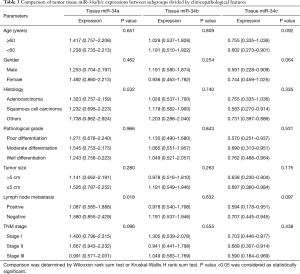
Full table
Association of plasma and tumor tissue miR-34a/b/c levels with DFS and OS
In the current study, we divided all patients into patients with miRNA-34a/b/c high expression and patients with miRNA-34a/b/c low expression subgroups based on their median value. Among these, plasma miR-34a high expression and miR-34a low expression were divided by the median value of 0.632, plasma miR-34b high expression and low expression were divided by the median value of 0.786, and plasma miR-34c high expression and low expression were divided by the median value of 0.342. As to tumor tissue, miR-34a high expression and low expression were divided by the median value of 1.323, tissue miR-34b high expression and low expression were divided by the median value of 0.786, and tissue miR-34c high expression and low expression were divided by the median value of 0.682. Kaplan-Meier curves suggested that plasma miR-34a (P=0.011, Figure 2A) and miR-34c (P=0.038, Figure 2B) high expression were correlated with prolonged DFS compared to low expression. Regarding tumor tissue, worse DFS was observed in patients with miR-34a (P=0.002, Figure 2C) low expression compared with high expression. Similarly, low miR-34c expression was correlated with shorter DFS (P=0.032, Figure 2D), while there was no difference of miR-34b high and low expression with DFS in plasma sample (P=0.302, Figure 2E) and tissue sample (P=0.134, Figure 2F).
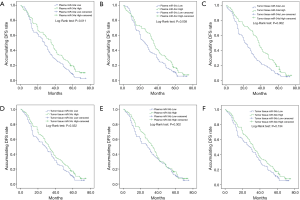
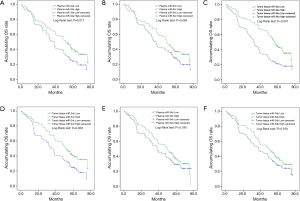
In addition, NSCLC patients with high expression of plasma miR-34a (P=0.011, Figure3A) or plasma miR-34c (P=0.064, Figure 3B) were observed in longer OS than that with low expression . Furthermore, tissue miR-34a (P<0.001, Figure 3C) and miR-34c (P=0.003, Figure 3D) higher expressions were correlated with longer OS. However, miR-34b high expression on OS was not different in low expression in both plasma sample (P=0.199, Figure 3E) and tissue sample (P=0.109, Figure 3F). Therefore, these results suggested that tumor tissue miR-34 expression was of better value in predicting prognosis compared with plasma expression, while the plasma miR-34a and miR-34c expression may be regarded as less invasive biomarkers for survival in some extent.
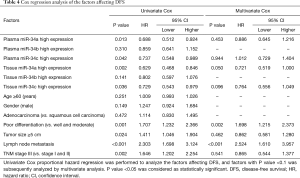
Cox regression analysis of the factors affecting DFS
All factors affecting DFS in NSCLC patients were analysed in univariate Cox proportional hazard regression, as presented in Table 4, which suggested that plasma miR-34a high expression (P=0.013), plasma miR-34c high expression (P=0.042), tissue miR-34a high expression (P=0.002) and tissue miR-34c high expression (P=0.036) were correlated with better DFS, while poor differentiation (P=0.001), tumor size ≥5 cm (0.024), lymph node metastasis (P≤0.001) and TNM stage (P=0.002) were negatively associate with DFS. Multivariate analysis was performed to further evaluate all factors with a P value <0.1 in univariate analysis. Tissue miR-34a high expression (P=0.050) was an independent predictive role in better DFS, while poor differentiation (P=0.002) and lymph node metastasis (P<0.001) could independently predict worse DFS in NSCLC patients.
Full table
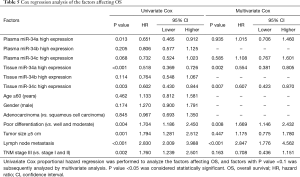
Cox regression analysis of the factors affecting OS
Univariate analysis was used to analyze the factors affecting OS in NSCLC patients, which disclosed that plasma miR-34a high expression (P=0.013), tissue miR-34a high expression (P<0.001) and tissue miR-34c high expression (P=0.003) were correlated with prolonged OS, while poor differentiation (P=0.004), tumor size ≥5 cm (P=0.001), lymph node metastasis (P<0.001) and TNM stage (P=0.002) were negatively associated with OS (Table 5). All factors with a P value <0.1 were subsequently evaluated with multivariate analysis. Tissue miR-34a high expression (P=0.002) and tissue miR-34c high expression (P=0.007) could independently predict better OS, while poor differentiation (P=0.008) and lymph node metastasis (P<0.001) could be independently risk factors of OS in patients with NSCLC.
Full table
Discussion
In the present study, we found that miR-34a/c expressions in the plasma were positively correlated with that in tumor tissue. Meanwhile, the plasma miR-34a expression and tumor tissue miR-34c expression were inversely associated with lymph node metastasis. Furthermore, miR-34a/c high expression in plasma and tumor tissue were both correlated with prolonged survival.
NSCLC, a heterogeneous disease with high morbidity and mortality (17), has been identified to arise from cancer cell proliferation and tumor capillaries development by multiple molecular abnormalities (18), such as anaplastic lymphoma kinase (ALK) gene abnormality through the produce of echinoderm microtubule associated protein like-4 gene-ALK (EML4-ALK) fusion transcript (19), and the epidermal growth factor receptor (EGFR) mutation, such as insertion mutations in exon 20 as well as exon 21, and so on (17,20). MiRNAs, as a single-stranded RNAs, inhibited the target genes and protein expression by binding to the 3'-untranslated regions (3'-UTRs) directly (21). A large amount of evidence has been identified that aberrant miRNAs expressions play critical roles in tumorigenesis, affecting the prognosis of cancer (6). For example, microRNA-520e regulate the target Zbtb7a-mediated Wnt signalling pathway, leading to the suppression of NSCLC cell growth (22). Additionally, endogenous miR-424 low expression serves as an effective repressor in NSCLC through the suppression of tumor cell proliferation and G1 to S phase cell-cycle transition (23). Therefore, several miRNAs have been identified to be associated with the pathologic process of NSCLC.
The homology of miR-34 family is located in 2–9 base pairs towards the 5' terminus of the mature miRNA sequence (12). Although miRNAs usually appear abnormal expression in tumor tissue sample, miRNAs expression in blood also dysregulated gradually, which could follow with blood circulation possibly due to tumor cell migration and invasion (24,25). Meanwhile, tissue biopsy, as a precise tool for diagnosis and tumor classification, still exists high risk of infection or haemorrhage by invasion, while the plasma sample, with non-invasions and easily obtained feature, is of more feasibility (26). Thus, our study extracted the plasma sample, and evaluated the correlation between miR-34a/b/c expressions in plasma and tumor tissues. The results were shown that miR-34a and miR-34c in plasma were positively associated with that in tumor tissues.
MiR-34a is sited in the second exon of a 33-kb transcript (EF570049), and has two promoter regions, including CpG island (CpGI) and p53-binding sites. Both of them downregulates miR-34 expression in cancers via hyper-methylation or controlling the stem cell compartment to inhibit malignancy respectively (27-29). Overexpression of miR-34a could contribute to tumor suppression via repressing human homolog of murine double minute 4 (HDM4) and p53 (30). Upregulated miR-34a expression could inhibit c-MET and AXL by extracellular signal regulated kinase (ERK) and serine/threonine kinase (AKT) signalling activation, suppressing diffuse malignant peritoneal mesothelioma (DMPM) cells proliferation and invasion, thereby resulting in the inhibition of tumorigenesis and tumor growth (31). Also, miR-34a could induce G1 cell cycle arrest, senescence and apoptosis to suppress tumor cells proliferation and promote differentiation, thereby repressing tumor growth and demonstrating the tumor-suppressive role (27); Furthermore, miR-34a overexpression could regulate the Snail1-mediated epithelial-mesenchymal transition (EMT) and the Notch signalling pathway to repress the migration and invasion of pancreatic cancer cells, thereby inhibiting tumor metastasis and pancreatic cancer progression (32). In terms of NSCLC patients, miR-34 family could regulate platelet-derived growth factor receptor alpha and beta (PDGFR-α and PDGFR-β) to promote tumor necrosis factor ligand superfamily member 10 (TRAIL)-induced apoptosis and reduce invasiveness of lung cancer cells, thereby inhibiting the migratory and invasive capacity of NSCLC cells (15). The results in our study were found that plasma miR-34a expression was negatively associated with lymph node metastasis, This might arise from the inhibition of cell proliferation, invasion as well as migration by miR-34a through regulating several pathways (27,30-32).
Previous study revealed that high miR-34a expression was associated with a lower risk of recurrence or death from breast cancer adjusted by multivariate analysis (33). Analogously, the downregulation of tumor tissue miR-34a was correlated with the poor prognosis of hepatocellular carcinoma (34). As for NSCLC, tumor tissue miR-34a was inversely correlated with recurrence-free survival (RFS) by regulation of c-MET and cyclin dependent kinase 6 (CDK 6) (35), while no similar study of plasma miR-34a in predicting prognosis of NSCLC is reported previously. Partially in line with this result, we found that miR-34a high expression was correlated with prolonged DFS and OS compared to low expression in this study, which partly resulted from its regulation of several pathways or genes, including c-MET and G1 phase regulators, to cause the inhibition of tumorigenesis and lymphatic invasion, thereby affecting recurrence of NSCLC patients (35). In addition, miR-34a increases the sensitivity of chemotherapy and radiotherapy, decreasing recurrence of NSCLC patients (36,37). Subsequently, Cox regression analysis was performed to evaluate factors affecting DFS and OS in this study, we also observed that plasma miR-34a was correlated with prolonged DFS and OS in univariate Cox model, while it could not independently predict DFS and OS of NSCLC patients in multivariate Cox model. This might result from that plasma miR-34a expression was correlated with its tissue expression and lymph node metastasis that contribute to the prognosis of NSCLC, which was identified before. Thus, plasma miR-34a expression might be affected by both of them, thereby influencing the predictive value of miR-34a in NSCLC patients. Furthermore, owning to the less invasion and more feasibility, plasma miR-34a was still a valuable biomarker to predict the prognosis in NSCLC patients.
MiR-34b/c, frequently responding on CpGI, are associated with B-cell translocation gene 4 (BTG4) (27). MiR-34c have been implicated to be a tumor suppressor of NSCLC via inhibiting pituitary adenylate cyclase-activating polypeptide type 1 (PAC1)/mitogen-activated protein kinase (MAPK) pathway, or retraining eukaryotic translation initiation factor 4E (eIF4E) expression (38,39). Down-expression of miR-34c was observed in lung tumor compared with that in normal tissues, which is resulted from their resistance to NSCLC cells migration and invasiveness by downregulating platelet-derived growth factor receptors (PDGFR-α/β) (15). Furthermore, increased expression of tissue miR-34c expression was associated with distant metastases formation in lung adenocarcinoma (LAC) via promoting hypermethylation (40). Similarly, our study observed that the plasma miR-34c high expression was correlated with prolonged DFS compared to low expression. The possible reasons might be the suppression of cells migration and invasiveness (15,38).
MiR-34b have been described to be epigenetically mediated in various cancer, such as colon cancer, head and neck cancer (41). Nevertheless, no relationship of miR-34b with DFS and OS both in plasma and tumor tissue was discovered in the present study. Prior research indicated the mediation of PDGFR-α and PDGFR-β, targeted by miR-34a/c, but not on miR-34b (15). Thus, one of the causes might be this.
Several limitations still existed in the present study. Firstly, the plasma miR-34 family expressions in healthy people were not assessed and compared with NSCLC patients, whereas many studies have reported decreased miR-34 expression in cancer patients compared with healthy controls group. Secondly, the detailed mechanism of miR-34 family in NSCLC was not analyzed. Thirdly, the analysis of lung cancer specific miRNAs was not explored, thus, further study about the investigation of lung cancer specific miRNAs in NSCLC patients is necessary. Finally, sample size was relatively small. The further study investigating the accurate mechanism of miR-34 family on the pathological process of NSCLC is needed.
In conclusion, circulating miR-34a and miR-34c might be served as novel prognostic biomarkers in NSCLC patients.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Ethics Committee of The Central Hospital of Wuhan, Tongji Medical College, Huazhong University of Science and Technology and written informed consent was obtained from all patients.
References
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [Crossref] [PubMed]
- Ginsberg MS, Grewal RK, Heelan RT. Lung cancer. Radiol Clin North Am 2007;45:21-43. [Crossref] [PubMed]
- Novaes FT, Cataneo DC, Ruiz RL Junior, et al. Lung cancer: histology, staging, treatment and survival. J Bras Pneumol 2008;34:595-600. [Crossref] [PubMed]
- Remark R, Becker C, Gomez JE, et al. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med 2015;191:377-90. [Crossref] [PubMed]
- Park SJ, More S, Murtuza A, et al. New Targets in Non-Small Cell Lung Cancer. Hematol Oncol Clin North Am 2017;31:113-29. [Crossref] [PubMed]
- Dong H, Lei J, Ding L, et al. MicroRNA: function, detection, and bioanalysis. Chem Rev 2013;113:6207-33. [Crossref] [PubMed]
- Du Y, Liu XH, Zhu HC, et al. MiR-543 Promotes Proliferation and Epithelial-Mesenchymal Transition in Prostate Cancer via Targeting RKIP. Cell Physiol Biochem 2017;41:1135-46. [Crossref] [PubMed]
- Xie F, Yuan Y, Xie L, et al. miRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Onco Targets Ther 2017;10:885-94. [Crossref] [PubMed]
- Zhao G, Wang B, Liu Y, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther 2013;12:2569-80. [Crossref] [PubMed]
- Li J, Chen Y, Guo X, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med 2017;21:838-47. [Crossref] [PubMed]
- Guo Y, Sun W, Gong T, et al. miR-30a radiosensitizes non-small cell lung cancer by targeting ATF1 that is involved in the phosphorylation of ATM. Oncol Rep 2017;37:1980-8. [Crossref] [PubMed]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 2010;17:193-9. [Crossref] [PubMed]
- Toyota M, Suzuki H, Sasaki Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res 2008;68:4123-32. [Crossref] [PubMed]
- Achari C, Winslow S, Ceder Y, et al. Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer 2014;14:538. [Crossref] [PubMed]
- Garofalo M, Jeon YJ, Nuovo GJ, et al. MiR-34a/c-Dependent PDGFR-α/β Downregulation Inhibits Tumorigenesis and Enhances TRAIL-Induced Apoptosis in Lung Cancer. PLoS One 2013;8:e67581. [Crossref] [PubMed]
- Tanaka N, Toyooka S, Soh J, et al. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer 2012;76:32-8. [Crossref] [PubMed]
- Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31. [Crossref] [PubMed]
- Fujimoto J, Wistuba II. Current concepts on the molecular pathology of non-small cell lung carcinoma. Semin Diagn Pathol 2014;31:306-13. [Crossref] [PubMed]
- Sullivan I, Planchard D. ALK inhibitors in non-small cell lung cancer: the latest evidence and developments. Ther Adv Med Oncol 2016;8:32-47. [Crossref] [PubMed]
- Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol 2005;23:2556-68. [Crossref] [PubMed]
- Bhatt K, Kato M, Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol 2016;310:F109-18. [PubMed]
- Zhijun Z, Jingkang H. MicroRNA-520e suppresses non-small-cell lung cancer cell growth by targeting Zbtb7a-mediated Wnt signaling pathway. Biochem Biophys Res Commun 2017;486:49-56. [Crossref] [PubMed]
- Wang Y, Lv Z, Fu J, et al. Endogenous microRNA-424 predicts clinical outcome and its inhibition acts as cancer suppressor in human non-small cell lung cancer. Biomed Pharmacother 2017;89:208-214. [Crossref] [PubMed]
- Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res 2011;39:7223-33. [Crossref] [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [Crossref] [PubMed]
- Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis 2014;5:e1327. [Crossref] [PubMed]
- Maroof H, Salajegheh A, Smith RA, et al. MicroRNA-34 family, mechanisms of action in cancer: a review. Curr Cancer Drug Targets 2014;14:737-51. [Crossref] [PubMed]
- Cheng CY, Hwang CI, Corney DC, et al. miR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Rep 2014;6:1000-7. [Crossref] [PubMed]
- Okada N, Lin CP, Ribeiro MC, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev 2014;28:438-50. [Crossref] [PubMed]
- El Bezawy R, De Cesare M, Pennati M, et al. Antitumor activity of miR-34a in peritoneal mesothelioma relies on c-MET and AXL inhibition: persistent activation of ERK and AKT signaling as a possible cytoprotective mechanism. J Hematol Oncol 2017;10:19. [Crossref] [PubMed]
- Tang Y, Tang Y, Cheng YS. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the Notch signaling pathway. Sci Rep 2017;7:38232. [Crossref] [PubMed]
- Peurala H, Greco D, Heikkinen T, et al. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS One 2011;6:e26122. [Crossref] [PubMed]
- Tian YW, Shen Q, Jiang QF, et al. Decreased levels of miR-34a and miR-217 act as predictor biomarkers of aggressive progression and poor prognosis in hepatocellular carcinoma. Minerva Med 2017;108:108-13. [PubMed]
- Hong JH, Roh KS, Suh SS, et al. The expression of microRNA-34a is inversely correlated with c-MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients. Tumour Biol 2015;36:9327-37. [Crossref] [PubMed]
- Pu Y, Zhao F, Li Y, et al. The miR-34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. BMC Cancer 2017;17:45. [Crossref] [PubMed]
- Lacombe J, Zenhausern F. Emergence of miR-34a in radiation therapy. Crit Rev Oncol Hematol 2017;109:69-78. [Crossref] [PubMed]
- Zhou YL, Xu YJ, Qiao CW. MiR-34c-3p suppresses the proliferation and invasion of non-small cell lung cancer (NSCLC) by inhibiting PAC1/MAPK pathway. Int J Clin Exp Pathol 2015;8:6312-22. [PubMed]
- Liu F, Wang X, Li J, et al. miR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif 2015;48:582-92. [Crossref] [PubMed]
- Daugaard I, Knudsen A, Kjeldsen TE, et al. The association between miR-34 dysregulation and distant metastases formation in lung adenocarcinoma. Exp Mol Pathol 2017;102:484-91. [Crossref] [PubMed]
- Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A 2008;105:13556-61. [Crossref] [PubMed]


