Exosomes as diagnostic and predictive biomarkers in lung cancer
Exosomes
Exosomes are small nanovesicles, in a range between 30 to 150 nm. Were first describe in 1983 in two paper published simultaneously (1,2). Exosomes are exocytosed by every cell in a constitutive manner, however, it has been demonstrated that tumour cells release higher amounts of exosomes than healthy ones and they can be found in different body fluids such as blood, semen, or ascites.
When discovered, exosomes were considered the garbage bags where the cells ousted the undesired components from themselves, however it has been demonstrated the implications of the exosomes in different functionalities of the tumour such as, immunomodulation (3), pre-metastatic niche formation (4), tumour growth (5), treatment resistance mediation (6) and more recently, exosomes mediated drug expulsion (7). Moreover, exosomes have been described to content messenger RNA (mRNA), microRNAs (miRNAs), double-stranded DNA (dsDNA) and proteins that could serve as diagnostic, predictive and prognostic biomarkers for the different tumours. More importantly, if exosomes are understood as horizontal cell communicators between cells, their content could mirror the one from the cell of origin; giving rise to a new and accessible source for tumour profiling analysis.
Extracellular vesicles such as exosomes accomplish a wide number of vesicles differing in size, origin and composition. Apoptotic bodies are big size vesicles ranging from 1,000 to 5,000 nm, smaller, macrovesicles can be found ranging from 200 until 1,000 nm, and in the small spectrum exosomes can be found ranging between 50 and 200 nm (8). However, due to their common multivesicular body (MVB) origin, all exosomes have a common profiling that allows their identification, based on different membrane proteins such as ALIX, TSG101 and CD63.
Different methods for exosomes isolation have been successfully used. The most common methods are ultracentrifugation and sucrose density-gradient ultracentrifugation or exosomes immunoprecipitation. However, recently new isolation methods have been used such as extracellular vesicle array or immunobeads precipitation (9,10). Both methods are based on antibodies recognition. Many other exosomes isolation kits have been developed recently based on filtration columns (11).
Exosomes biogenesis and trafficking
Originally, exosomes were thought to be released directly through fusion with the plasmatic membrane. However, was not until the 80s when it was described the existence of an intracellular endosome, that leaded to the formation of an MVB, that later, could release exosomes through fusion with the plasma membrane (2).
The endosomal sorting complex required for transport (ESCRT) was firstly described to control the exosomes formation inside the MVB. It consists in four main complexes with other auxiliary proteins. The ESCRT-0 controls the cargo clustering in a ubiquitin dependent manner. The ESCRT-I and II induce the bud formation in the MVB and the ESCRT-III induce the vesicle scission from the MVB membrane. The auxiliary VPS4 protein is in charge of recycling the ESCRT machinery (12) (Figure 1). Until recently, the ESCRT mechanism was the only known to induce the exosomes formation, however, in the last years; many articles have been published demonstrating alternative pathways for the exosomes formation ESCRT-independent. These mechanisms include proteins known to be present in the exosomes. The cholesterol has been described to induce the vesicles secretion in Flotillin-2 dependent manner (13). The phospholipase D2 (PLD2), increase the production of the intraluminal vesicles in the MVB through the increase of the inward curvature of the MVB membrane (14). Moreover, different tetraspanins have been demonstrated to have any active role in the mRNA and protein cargo sorting in the exosomes (15,16).
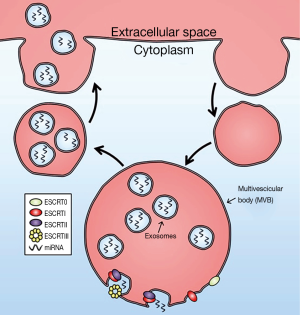
Once the exosomes are in the luminal space of the MVBs, SNARE family, and concretely, VAMP-7 could be enhancing the membrane fusion between the MVBs and the plasma membrane. However, the inhibition of the proteins from the SNARE family does not result in the complete depletion of exosomes secretion, suggesting other proteins could play this role (17,18). Finally, the exosomes secretion is mostly induced by proteins from the RAB family such as RAB27a and RAB27b (19).
Exosomes as markers for non-small cell lung cancer (NSCLC)
One of the most attractive aspects of exosomes research is the discovery of novel biomarkers for the early and improved detection of lung cancer (20). The content of the exosomes has been widely described in different tumours. miRNAs are probably the most studied molecules inside the exosomes. Among miRNAs, miRNA-21 have been widely described in NSCLC exosomes, and its impact in other malignancies has been described in several reports related with tumour growth in both haematological and solid malignancies, for example, in diffuse large B-cell lymphoma the AKT signalling pathway is constitutively activated due to the over expression of miR-21 that leads to the suppression of forkhead box protein O1 expression and the downregulation of PTEN expression, leading to a poor prognosis of the disease (21,22). Regarding the solid tumours, breast cancer and colorectal cancer have been demonstrated to upregulate miR-21 in advanced stages leading to a poor prognosis of the disease and high risk of metastasis (23,24). Also, mRNA have been described in the exosomes also as a possible predictive biomarker that could help to improve, in a non-invasive way, the treatment choice and to predict the resistance before clinical significances appear (25). Many proteomic studies have been performed in different tumours inside the exosomes with successfully results, being able to create protein profiles with diagnostic and prognostic functions, and also to identify exosomal proteins that can play key roles in the receptor cells (25,26). In the last years and since the discovery of the exosomal dsDNA in 2014 (27), the studies on the field has increased exponentially.
Many correlations have been found between tissue biomarkers and exosomal biomarkers in different tumours opening the door to improve our detection of diagnostic, predictive and prognostic biomarkers. Although none has been yet validated for NSCLC analysis, in the following paragraphs the main advances and most promising markers will be described. A summary of all the markers described can be found in Table 1.
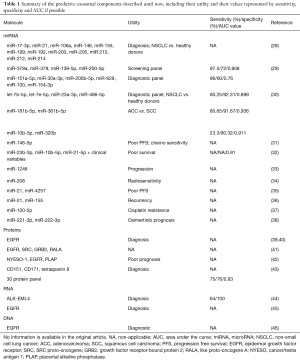
Full table
miRNA
miRNAs are small non-coding RNAs. They are recognized to regulate a number of genes by binding to the 3’UTR of their target mRNA, resulting in the alteration of the targeted gene expression. A single miRNA can influence multiple genes in a single cell. The utility of miRNAs for cancer study has been largely described. Unlike circulating miRNAs, exosomal miRNAs are protected from the degradation by RNAse in the blood torrent (47), thus miRNAs could serve as a useful tool in three fields of study; diagnostic of NSCLC, prognosis against a treatment and risk of resistance after a therapy.
The first group of miRNAs was described to be useful to predict NSCLC over healthy patients by Rabinowits et al. in 2009, it consists of 12 miRNAs, miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-199, miR-192, miR-203, miR-205, miR-210, miR-212 and miR-214, which were present in NSCLC both in tissue and in circulating exosomes being their levels non-detectable in healthy donors (28). In the same paper, they described that the miRNA levels inside the exosomes derived from NSCLC were higher compared to those from healthy donors (28). Cazzoli et al. were the first groups to develop 2 miRNA profiling, one for screening based on 4 miRNAs, miR-378a, miR-379, miR-139-5p and miR-200-5p, and one for diagnosis based on 6, miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100 and miR-154-3p, both extracted from a wide analysis of 746 miRNAs analysed on a training set of 3 groups: lung adenocarcinomas, lung granulomas, healthy former smokers. These profiles were confirmed in a validation set of 50 patients. In the case of the screening test the sensitivity was 97.5% for 72% specificity and an area under the curve (AUC) value of 0.908 while the diagnostic test exhibited a sensitivity of 96% for 60% of specificity with an AUC value of 0.76 (29). More recently, Jin et al. has developed a miRNA profile based on 4 miRNAs, let-7b-5p, let-7e-5p, miR-23a-3p, and miR-486-5p that exhibited sensitivity of 80.25% and a specificity of 92.31% with an AUC value of 0.899 being able to diagnose 43 NSCLC over 60 samples. In the same paper, levels of miR-181b-5p and miR-361b-5p, as well as miR-10b-5p and miR-320b were used to identify among the 43 patients 31 adenocarcinoma patients and 11 squamous cell carcinoma (SCC) patients respectively. The miRNA for adenocarcinoma and SCC diagnosis showed an AUC value of 0.936 and 0.911 respectively with a sensitivity of 80.65% and a specificity of 91.67% for adenocarcinoma and a sensitivity of 83.33% and a specificity of 90.32% for SCC (30).
One of the big problems of NSCLC is the high rates of progression and recurrence of the patients; thus, tools to predict this behaviour are needed to improve the medical treatment choices in the clinic. Many miRNAs have been reported to be differentially expressed in situations where the tumour is progressing before it is a detectable under computerize tomography (CT) scan.
The downregulation of the miRNA-146-5p is indicative of a poor progression free survival (PFS) compared to those patients with higher levels of the miRNA inside the exosomes. Moreover, its levels are correlated to the chemosensitivity of the tumour to Cisplatin mediated through the blockade expression of Atg12, an autophagy mediator gene (31). In another study with 10 lung cancer patients and 10 healthy controls, 83 cancer-related miRNAs were analysed. After informatics analysis, 9 miRNAs were differentially expressed and validated in a subset of 209 NSCLC patients. From those, miR-23b-3p, miR-10b-5p and miR-21-5p were validated as upregulated miRNAs in NSCLC compared to healthy controls. Separately, the 3 miRNAs showed significantly low overall survival in those patients with higher levels of the miRNA in their exosomes compared to those with low levels. Moreover, the 3 miRNAs model improves the AUC value from 0.88, from only the clinical variables, to 0.91 analysing both the miRNAs expression and the clinical variables such as sex, or tobacco consumption (32). Exosomal miRNA has been described also to play a role in the predictive response to a treatment as it has been explained before with the miRNA-146-5p (31). Two other independent studies show that upregulation of miR-1246 and miR-208-a is correlated with a high proliferation of the tumour and a resistant profile to radiotherapy by targeting the genes DR5 and p21 respectively. This could lead not only to a prognostic biomarker but to a new target against NSCLC (33,34). The miR-21 and miR-4257 were found among a subset of patients with recurrence after surgery, without recurrence and healthy donors. These results were validated in 201 cases with an upregulation of 2.5- and 2.9-fold of increase respectively between recurrence patients and healthy donors. Moreover, it was demonstrated that the disease-free survival-similar to PFS, was lower in those patients with high expression of both miRNAs considered high more than the mean of the expressions. They described also the correlation of the over expression of miR-21 with the tumour size and TNM stage, and the correlation between de levels of miR-4257 and lymphatic node invasion, histological type and TNM stage (35).
One of the problems of the research and analysis of exosomes is the amount of material available. In another study, exosomes isolated from H1299 (NSCLC cell line) and Beas-2b (bronchial-epithelial cell line), and isolated from mouse with H1299 after surgery, some being recurrent and some not. The authors faced a very low-positive amplification rate due to the low amount of extracted RNA, and high variability between replicates and groups. However, despite the low amplification rates, the authors showed that miR-21 and miR-155 may be upregulated in the group of mice recurrent after surgery in comparison with primary tumour mice (36).
One of the most significant papers regarding the utility of miRNA inside the exosomes has been recently published by Qin et al. They described that miR-100-5p is downregulated in exosomes derived from A549 resistant to Cisplatin (A549/DDP) in comparison with A549 wild-type. However, they saw that exosomes derived from A549/DDP were able to transfer Cisplatin resistance mediated by the binding of miR-100-5p to the 3’ UTR of mTOR gene. Same experiments were performed in vivo with mice inoculated in different conditions of exosomes derived from A549, A549/DDP and inhibitors and mimics of miR-100-5p being the mice with exosomes of A549/DDP transfected with miR-100-5p control mimic they group with a higher tumour growth, lower levels of miR-100-5p and higher levels of mTOR (37).
Contrary to what it has been shown in this review, some papers have demonstrated a negative correlation between tissue and exosomal miRNA levels. Our group published an abstract with a follow up analysis of patients during Osimertinib treatment, a third-line tyrosine kinase inhibitor. The treatment lead to an upregulation of oncomiRs (hsa-miR-221-3p/222-3p) in exosomes isolated from patient’s plasma respect to healthy control. However, the upregulation was correlated to a good clinical outcome suggesting that some these two miRNAs may be pump out of the cell due to the treatment (38).
All these studies suggest that exosomal miRNA varies their composition depending on their cellular origin being able to differentiate between NSCLC subtypes, and due to their stability and the stability of their content they may be perfect candidates for the diagnosis, or prognosis of NSCLC (29-31,36).
Exosomal protein
Exosomes can be considered as potential tools for tumour diagnosis and prognosis. These stable vesicles, contained in the blood stream, are tumour specific and protected from degradation by a lipid bilayer membrane. Recent studies identify the proteome of cancer exosomes; proteomic analyses show that several proteins are enriched in exosomes. These proteins are involved in exosome biogenesis, membrane transport and fusion and in various steps of tumour progression such as metastasis, angiogenesis and immunomodulation (48)
According to the database Exocarta (http://www.exocarta.org), 9,769 proteins have been identified in exosomes. Some exosomal proteins can mirror producer cells and pathological stage of disease (48). Moreover, some of these proteins play a key role in tumorigenesis. These features qualify exosomal protein as good diagnostic and prognostic potential biomarkers in lung cancer (49).
Lung cancer exosomes contain several tumours associated proteins, such as epidermal growth factor receptor (EGFR), KRAS, extracellular matrix metalloproteinase inducer (EMMPRIN), claudins and RAB-family proteins. CD91, CD317 and EGFR have been suggested as potential exosomal markers in NSCLC. Exosomal EGFR is one membrane-bound proteins that have been evaluated in NSCLC (39). Specifically, Huang et al. found that 80% of the exosomes isolated from NSCLC biopsies were EGFR positive (40).
Recently, it was tested exosomal proteins differential expressed in normal bronchial epithelial cells and NSCLC cells using a triple SILAC quantitative proteomic method. They found that NSCLC exosomes are enriched in proteins involved in cell signalling, cell adhesion extracellular matrix remodelling. They identify and quantify 721 exosomal proteins derived from three cell lines. Among the proteins associated with signal transduction, enriched in NSCLC exosomes, EGFR and SRC are upregulated as well as downstream effectors such as growth factor receptor-bound protein 2 (GRB2) and RALA. Moreover, it was demonstrated that MET receptor, RAC1, and KRAS proteins were increased in NSCLC exosomes (41).
Furthermore, Sandfeld-Paulsen demonstrated that in a cohort of 276 non-selected NSCLC patients of all stages, it was evaluated 49 exosomal membrane bound proteins and found that nine proteins have a potential as prognostic marker in NSCLC. In particular, they indicate that increasing concentration level of NYESO-1, EGFR and PLAP are prognostic markers of poor prognosis (42). Proteomic analysis of exosomes isolated from human malignant pleural effusions demonstrated that these vesicles contained MHC class I and II proteins, heat shock and cytoskeletal proteins, and signal transduction—involved proteins. These proteins have already been reported as constituent of exosomes from other origin.
Exosomes from malignant pleural effusions contain peculiar proteins, such as Sorting-nexin (SNX) family, a group of hydrophilic proteins involved in the intracellular trafficking of proteins to different organelles. Acidic ribosomal phosphoproteins, from the 60S subunit of ribosomes, interact with elongation factors EF-1 and EF-2 and play an important role in the elongation step of protein synthesis. The presence in exosomes of proteins involved translation in may also be explained by their high concentration in the cytosol of cancer cells (50)
Nowadays, in the field of the biomarker discovery, the methodology that focuses only on a single exosome-protein has been overtaken that multiple protein markers panel (8). In particular, Jakobsen and colleagues used an extracellular vesicle array with 37 antibodies to capture exosomes directly from the plasma of NSCLC patients, the result indicated that the sensitivity, specificity and diagnostic accuracy of combined 30-marker model (43). A recent study used the extracellular vesicle array with 49 antibodies. Among the 49 exosomal proteins, CD151, CD171, and tetraspanin 8, have been considered the strongest markers to identify lung cancer of all histological subtypes from the control. It could important not only to evaluate protein profiling of exosomes from different lung cancer stage and histology, but also as potential diagnostic tool for lung cancer (51).
RNA and DNA
Due to the difficulties working with exosomes, it has not been until recent years with the improvement in the sensitivity of the techniques and the discovery of the high throughput technologies, such as NGS, that exosomal RNA and DNA may become a real alternative to the tissue analysis.
Our group has recently described for the first time the anaplastic lymphoma kinase (ALK)-EML4 translocation inside the exosomes with a sensitivity of 64% and a specificity of 100% over a subset of 17 patients being 14 positive and 3 negative compared with tissue from the same patients (44). Just few months earlier, Krug et al. detected for the first time RNA transcribed from the EGFR gene, in a study comparing the analysis of ctDNA for EGFR mutation with a combined analysis of ctDNA together with exosomal RNA presenting a better sensitivity for all the analysed mutations (45).
In 2014, Thakur et al. described for the first time the presence, inside chronic leukaemia, human colorectal carcinoma, and murine melanoma derived exosomes (DEX) of dsDNA over other forms of DNA. However, the presence of exoDNA inside the lung cancer DEX was lower than the in other tumours. To conclude the research, the authors performed an analysis of exosomal EGFR in 4 NSCLC cell line with the objective of detecting mutations known to be present in those cell lines. The results showed that 100% of the exoDNA analysis were positive for the corresponding mutation (46).
ExoDNA may lead to improvements in the sensitivity and specificity of detecting tumour alterations that will prove useful in the clinical setting in the analysis of driver mutations (52). Moreover, their stability and abundance makes them very good candidates over other liquid biopsy compartments exposed constantly to the conditions of the blood stream.
Discussion and future perspectives
Exosomes is a relatively new field of study; thus, no established protocols are available for their isolation or analysis. However, and despite the lack of consensus about how to work with exosomes, many groups are publishing interesting researches on the field. These facts demonstrate that although there is still a long way to run in the understanding of exosomes and their functions, exosomes can play a role not only in the diagnosis and prognosis of cancer, but can help us to understand the mechanisms of the cancer, to elucidate new targets even to function as specific drug deliverers.
We have already written about the miR-100-5p described by Qin et al. In this case, miRNA does not play only a function of prognosis biomarker but induces a change in the receptor cell producing an increase in the resistance to cisplatin (37). Similar results were described by Liu et al. They saw that the upregulation of the gene TLR3 increase the cytokine production leading to pre-metastatic niche formation and metastasis. However, they saw that the activation of TLR3 in the lung epithelial cells was produced by exosomes derived from the tumour, and concretely, by different U1 snRNA presented in very high concentrations, compared to non-tumoral DEX (53). This means, that exosomes do actually play a function in the tumour progression and metastasis leading to new molecules that can be targeted in order to stop the tumour development.
Exosomes mirror the lipid bilayer present in the cells due to their exocytic origin, and, due to the same reason, they would be able to carry surface proteins and fuse with the membrane of the receptor cell to release their cargo, and due to their stability, they would be able to carry molecules of interest without risk of alteration or degradation (54). Based on this idea a Phase I immunotherapeutic study was carried on using Dendritic cell DEX loaded with Melanoma associated antigen (MAGE), the results showed a good NK reactivity with low side effects (55). In another study, it was analysed the effect of a second generation DEX coupled with chemotherapy showing a better activation of T cells compared to normal DEX resulting on a better control of the tumour (56). Very few research have been done in this field, but due to the autology on the use of DEX, it is easy to think that the side effects derived from an adjuvant therapy based on exosomes will not exceed the side effects of the principal therapy.
In conclusion, exosomes are still a new field of study, where a consensus is needed to be able to include protocols in the clinical practice as it is already established for Erlotinib and Osimertinib in EGFR mutation detected in cfDNA. Very few is still known about how to use exosomes as adjuvants in a therapy or as drug deliverers, however, the new technologies and investigations are bringing these advances closer in time. Also, there is a lack of information about how exosomes interact with other tissues in longer distances making difficult the understanding of their function in the tumour progression and metastasis. This current review has focused on how the exosomes can be the future of biomarkers identification for diagnostic and prognostic of NSCLC specially miRNA and proteins, the most studied. A deeper study of exosomes and its composition will lead to better biomarker identification and thus, an early detection of the tumour.
Acknowledgements
Elena Durendez has a predoctoral fellowship by Asociacion Española Contra el Cancer (AECC, Valencia, Spain) and Paolo Manca for his contribution creating the figure for this review.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 1983;33:967-78. [Crossref] [PubMed]
- Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 1983;97:329-39. [Crossref] [PubMed]
- Chow A, Zhou W, Liu L, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci Rep 2014;4:5750. [Crossref] [PubMed]
- Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015;17:816-26. [Crossref] [PubMed]
- Raimondo S, Saieva L, Corrado C, et al. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun Signal 2015;13:8. [Crossref] [PubMed]
- Hupfeld T, Chapuy B, Schrader V, et al. Tyrosinekinase inhibition facilitates cooperation of transcription factor SALL4 and ABC transporter A3 towards intrinsic CML cell drug resistance. Br J Haematol 2013;161:204-13. [Crossref] [PubMed]
- Safaei R, Larson BJ, Cheng TC, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther 2005;4:1595-604. [Crossref] [PubMed]
- Whiteside TL. Tumor-Derived Exosomes and Their Role in Tumor-Induced Immune Suppression. Vaccines (Basel) 2016;4:E35. [Crossref] [PubMed]
- Jørgensen M, Bæk R, Pedersen S, et al. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles 2013.2. [PubMed]
- Zarovni N, Corrado A, Guazzi P, et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015;87:46-58. [Crossref] [PubMed]
- Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics 2017;7:789-804. [Crossref] [PubMed]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25. [Crossref] [PubMed]
- Strauss K, Goebel C, Runz H, et al. Exosome secretion ameliorates lysosomal storage of cholesterol in Niemann-Pick type C disease. J Biol Chem 2010;285:26279-88. [Crossref] [PubMed]
- Ghossoub R, Lembo F, Rubio A, et al. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat Commun 2014;5:3477. [Crossref] [PubMed]
- van Niel G, Charrin S, Simoes S, et al. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 2011;21:708-21. [Crossref] [PubMed]
- Nazarenko I, Rana S, Baumann A, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res 2010;70:1668-78. [Crossref] [PubMed]
- Fader CM, Sánchez DG, Mestre MB, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta 2009;1793:1901-16. [Crossref] [PubMed]
- Rao SK, Huynh C, Proux-Gillardeaux V, et al. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J Biol Chem 2004;279:20471-9. [Crossref] [PubMed]
- Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 2010;12:19-30; sup pp 1-13.
- Alipoor SD, Mortaz E, Garssen J, et al. Exosomes and Exosomal miRNA in Respiratory Diseases. Mediators Inflamm 2016;2016:5628404. [PubMed]
- Go H, Jang JY, Kim PJ, et al. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget 2015;6:15035-49. [Crossref] [PubMed]
- Hong JY, Hong ME, Choi MK, et al. The impact of activated p-AKT expression on clinical outcomes in diffuse large B-cell lymphoma: a clinicopathological study of 262 cases. Ann Oncol 2014;25:182-8. [Crossref] [PubMed]
- Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 2008;14:2348-60. [Crossref] [PubMed]
- Nielsen BS, Jorgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis 2011;28:27-38. [Crossref] [PubMed]
- Jenjaroenpun P, Kremenska Y, Nair VM, et al. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ 2013;1:e201. [Crossref] [PubMed]
- Silva J, Garcia V, Rodriguez M, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer 2012;51:409-18. [Crossref] [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. [Crossref] [PubMed]
- Rabinowits G, Gerçel-Taylor C, Day JM, et al. Exosomal MicroRNA: A Diagnostic Marker for Lung Cancer. Clin Lung Cancer 2009;10:42-6. [Crossref] [PubMed]
- Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. [Crossref] [PubMed]
- Jin X, Chen Y, Chen H, et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin Cancer Res 2017;23:5311-9. [Crossref] [PubMed]
- Yuwen DL, Sheng BB, Liu J, et al. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2017;21:2650-8. [PubMed]
- Liu Q, Yu Z, Yuan S, et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017;8:13048-58. [PubMed]
- Yuan D, Xu J, Wang J, et al. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 2016;7:32707-22. [Crossref] [PubMed]
- Tang Y, Cui Y, Li Z, et al. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res 2016;35:20. [Crossref] [PubMed]
- Dejima H, Iinuma H, Kanaoka R, et al. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett 2017;13:1256-63. [PubMed]
- Munagala R, Aqil F, Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol 2016;37:10703-14. [Crossref] [PubMed]
- Qin X, Yu S, Zhou L, et al. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int J Nanomedicine 2017;12:3721-33. [Crossref] [PubMed]
- Giallombardo M, Jorge Chacartegui J, Reclusa P, et al. Follow up analysis by exosomal miRNAs in EGFR mutated non-small cell lung cancer (NSCLC) patients during osimertinib (AZD9291) treatment: A potential prognostic biomarker tool. Available online: http://meeting.ascopubs.org/cgi/content/abstract/34/15_suppl/e23035
- Yamashita T, Kamada H, Kanasaki S, et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013;68:969-73. [PubMed]
- Huang SH, Li Y, Zhang J, et al. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest 2013;31:330-5. [Crossref] [PubMed]
- Clark DJ, Fondrie WE, Yang A, et al. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J Proteomics 2016;133:161-9. [Crossref] [PubMed]
- Sandfeld-Paulsen B, Aggerholm-Pedersen N, Baek R, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol 2016;10:1595-602. [Crossref] [PubMed]
- Melton L. Protein arrays: proteomics in multiplex. Nature 2004;429:101-7. [Crossref] [PubMed]
- Rolfo C, Laes JF, Reclusa P, et al. P2.01-093 Exo-ALK Proof of Concept: Exosomal Analysis of ALK Alterations in Advanced NSCLC Patients. J Thorac Oncol 2017;12:S844-5. [Crossref]
- Krug AK, Karlovich C, Koestler T, et al. Abstract B136: Plasma EGFR mutation detection using a combined exosomal RNA and circulating tumor DNA approach in patients with acquired resistance to first-generation EGFR-TKIs. Am Assoc Cancer Res 2016;14:B136.
- Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9. [Crossref] [PubMed]
- Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012;7:e30679. [Crossref] [PubMed]
- Zhou L, Lv T, Zhang Q, et al. The biology, function and clinical implications of exosomes in lung cancer. Cancer Lett 2017;407:84-92. [Crossref] [PubMed]
- Mirzaei H, Masoudifar A, Sahebkar A, et al. MicroRNA: A novel target of curcumin in cancer therapy. J Cell Physiol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Bard MP, Hegmans JP, Hemmes A, et al. Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am J Respir Cell Mol Biol 2004;31:114-21. [Crossref] [PubMed]
- Jakobsen KR, Paulsen BS, Baek R, et al. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles 2015;4:26659. [Crossref] [PubMed]
- Lyden DD. Tests for Double-Stranded DNA Fragments in Exosomes Could Lead to Earlier, Easier Cancer Diagnoses and Targeted Treatments. Available online: http://news.weill.cornell.edu/news/2014/05/tests-for-double-stranded-dna-fragments-in-exosomes-could-lead-to-earlier-easier-cancer-diagnoses-an
- Liu Y, Gu Y, Han Y, et al. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell 2016;30:243-56. [Crossref] [PubMed]
- Johnsen KB, Gudbergsson JM, Skov MN, et al. A comprehensive overview of exosomes as drug delivery vehicles - endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta 2014;1846:75-87. [PubMed]
- Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med 2005;3:9. [Crossref] [PubMed]
- Taieb J, Chaput N, Novault S, et al. Dendritic cell derived-exosomes boost effector CD8+ T cells in the absence of tumor induced-T regulatory cells: Synergistic antitumor effects of cyclophosphamide and exosome-based vaccines. Available online: http://cancerres.aacrjournals.org/content/65/9_Supplement/623.4.abstract


