Respecting boundaries: CTCF, chromatin structural organization, and heart failure
The mammalian heart is highly adaptable, responding to growth of the organism, and to the rapidly changing demands to deliver nutrients and energy throughout the body based on exercise or rest, feeding or fasting, infection, injury and other stresses. In addition to acute changes in cardiac function to meet immediate demands, the heart has an impressive capacity for remodeling in response to long-term challenges, such as the physiological hypertrophy that occurs in the athlete’s heart, as well as the pathological hypertrophy due to chronic hypertension (1). Cardiac hypertrophy is initially an adaptive response to this pressure overload, but over time this remodeling becomes maladaptive, leading to inefficient contractions characteristic of heart failure. Heart failure affects over 23 million people worldwide and is a major cause of death (2,3). There are currently limited treatments for severe heart failure, including left ventricular assist devices (LVAD), which are implanted to enhance heart function, often as a bridge to full cardiac transplantation. In light of this problem, new understanding of heart failure is urgently warranted. Rosa-Garrido et al. (4) provide new insights into the relationship between chromatin organization in the heart and gene expression, heart function, and failure.
Chromatin architecture and organization in the nucleus is a tightly regulated process that is essential for normal biological function. DNA is wrapped around histone protein complexes to form nucleosomes (Figure 1). These fold into higher order structures of chromatin which allow for compaction of the ~2 meters of DNA within a single human cell nucleus. Chromatin organization also allows for controlled accessibility to gene regulatory elements. Oftentimes, these regulatory elements such as enhancers and promoters are separated by relatively long distances along linear DNA (5), but chromatin looping allows for interactions between these long-range regulatory domains. Chromatin is also organized into higher order domains known as topologically associating domains (TADs). These are regions of chromatin that physically interact with each other with high frequency in a regulated manner, allowing for coordinated control of gene expression and timing of DNA replication (5). Studies have increasingly revealed a cell type-specific organization of chromatin within the nucleus, with actively transcribed genes more localized to the nuclear interior region, while silenced/inactive genes are most often located near the nuclear periphery (6). Although the spatial organization of chromatin within the nucleus is clearly non-random and therefore likely important, the mechanisms governing this positioning are not well understood.
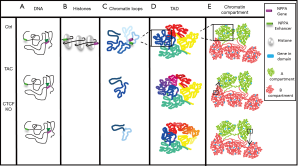
Two protein factors play a primary role in formation and stabilization of chromatin loops and TADs: cohesin and CCCTC-binding factor (CTCF). Cohesin is a ring-like multiprotein complex that plays a role in cohesion of sister chromatids during DNA replication and cell division and maintains long-range interactions between cis-regulatory elements (7). Cohesin along with CTCF, a multi-functional DNA binding protein, co-occupy thousands of genomic binding sites (8). CTCF is an architectural protein with many diverse functions. It is the main insulator protein in vertebrates and can prevent interaction between enhancers and promoters (9). CTCF bindings sites are enriched at TAD boundaries and deletion of CTCF or its binding sites at these boundaries influences inter- and intra-domain interactions and, subsequently, regulation of gene expression (5,9). Investigation of the roles of CTCF and related factors in regulation of chromatin looping and maintenance of higher order chromatin structure has been aided by the advent of techniques such as chromatin conformation capture-based approaches.
Hi-C is a chromatin conformation capture, or 3C, method that takes advantage of proximity ligation to allow for investigation of 3D genome organization (10). In Hi-C, cellular DNA is crosslinked and fragmented with restriction enzymes followed by ligation of adjacent DNA fragments (5). Subsequently, crosslinking is reversed and fragments are biotinylated. These fragments can then be pulled down using streptavidin and genome-wide interactions can be identified by high-throughput sequencing. These 3C-based methods allow for a more thorough understanding of the basics of 3D chromatin organization and have revealed its dysregulation in diseases such as cancer and limb malformation (10). While little is known about how global chromatin organization in the heart affects function, an initial study using 3C methods demonstrated that CTCF deletion in cardiac progenitor cells in vivo alters local chromatin interactions, leading to changes in gene expression that promote cardiac defects and embryonic lethality (11). However, the global chromatin structure in adult cardiomyocytes and the changes that take place in heart failure are unknown.
Rosa-Garrido and colleagues recently employed 3C technology to map for the first time the 3D chromatin organization in adult cardiomyocytes. In this study, they also assessed how pressure overload-induced hypertrophy or CTCF loss of function influence 3D chromatin organization and heart function (4). They report that an inducible cardiomyocyte-specific knockdown of CTCF (referred to as CTCF-KO) induces cardiomyopathy as measured by decreased ejection fraction, increased left ventricular internal diameter and fibrosis, and cardiac hypertrophy. Notably, this dramatic phenotype was apparent with an 80% reduction in CTCF expression in myocytes, and any further reduction in CTCF expression resulted in death. While the CTCF-KO phenotype shared some features of pressure overload-induced cardiac remodeling in response to transverse aortic constriction (TAC), the CTCF-KO phenotype was more severe and some of these responses were distinct suggesting pathologically different mechanisms underlying cardiac remodeling in these models. Interestingly, expression of CTCF changes in some instances of cardiac remodeling. The authors found that TAC did not alter CTCF protein levels. However, CTCF was downregulated in isoproterenol-induced hypertrophy in mice, but upregulated in heart failure patients after treatment with LVAD, which reduces load on the heart. These findings reveal that disruption of CTCF function leads to changes in chromatin organization and gene expression that can cause hypertrophic cardiomyopathy, and that changes in CTCF expression, likely resulting in chromatin reorganization, can also be associated with some stimuli of cardiac remodeling.
The differences in hypertrophic pathology between TAC and CTCF-KO mice were also reflected in the chromatin architecture and differential changes in gene expression in the cardiomyocytes of these mice. Although the number of TAD boundaries and compartmentalization of chromatin in CTCF-KO and TAC mice didn’t change as compared to control mice, the boundary strength was altered across the genome. The accompanying transcriptional analyses revealed less than half of the differentially regulated genes were shared between TAC and CTCF-KO mice compared with controls. Furthermore, although total long-range looping of chromatin was decreased in both CTCF-KO and TAC mice, only half of the disappearing loops and 15% of the appearing loops were shared between the two groups of mice. A subset of the loops that were altered in CTCF-KO and TAC mice are involved in bringing together enhancers and promoters.
To identify how structural changes in chromatin alter gene expression in heart failure, the authors investigated local chromatin structure in CTCF-KO and TAC mice. They found that there were decreased intra-chromosomal interactions in genes associated with heart failure such as Nppa and Mef2c. Most of the interactions of genes expressed (60%) in CTCF-KO and TAC mice changed in a similar direction. Of the genes that had interactions in CTCF-KO and TAC mice that were differentially expressed compared to control, the majority of the gene expression changes were a result of decreased chromatin associations. Moreover, enhancers also displayed decreased interactions in the cardiomyocytes of CTCF-KO and TAC mice. These findings demonstrate that heart failure results in more dynamic global changes in chromatin architecture and local changes in gene and enhancer interaction.
The present study is the first to describe TAD organization, compartmentalization, chromatin looping, and remodeling of enhancers and regions surrounding genes in adult cardiomyocytes and how these elements are restructured in disease. Moreover, it provides further evidence of the complexity of gene expression which extends beyond transcriptional regulation to include modifications to DNA and histones, long distance interaction of regulatory elements, and global changes in chromatin structure. Together, these multiple layers of regulation likely underlie pathogenesis of numerous diseases and their cooperation should be considered in conditions in addition to heart failure. CTCF is known to play an important role in regulation of chromatin conformation (7,12), and the same group has previously reported that CTCF works mutually with another structural protein, Hmgb2, to regulate gene expression in cardiomyocytes (13). In this study, Rose-Garrido and colleagues extended these and their previous findings to provide the first description of changes in global chromatin architecture and a cardiomyopathy phenotype following cardiomyocyte-specific CTCF knockout in adult mice.
Although this study advances our understanding of chromatin conformation changes that occur in a TAD-induced model of heart failure and following CTCF-knockout, it highlights several questions to be investigated: (I) identify whether the changes in chromatin architecture are a cause or consequence of alterations in cardiomyocytes that occur in cardiomyopathies and other causes of heart failure; (II) determine what causes dysregulation of chromatin organization in heart failure; (III) better understand the role of CTCF and other chromatin binding proteins in the pathogenesis of heart failure; and (IV) characterize changes in chromatin structure that occur during development including determining how structural proteins, chromatin loops, and TADs are initially organized. It will be of particular interest to determine how expression of cardiac master regulatory transcription factors and epigenetic regulators are spatially regulated and how this is altered in response to hypertrophic stimuli. Answering these questions will allow us to better understand how these processes may fail in diseases like heart failure.
Acknowledgements
Funding: The authors thank the National Science Foundation (DGE-1122492 to AJ Sizer) and the National Institutes of Health (HL091013, HL118430, RHL119529 to KA Martin) for funding.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Shimizu I, Minamino T. Physiological and pathological cardiac hypertrophy. J Mol Cell Cardiol 2016;97:245-62. [Crossref] [PubMed]
- Writing Group Members, Mozaffarian D, Benjamin EJ, et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation 2016;133:e38-360. [Crossref] [PubMed]
- Roger VL. Epidemiology of Heart Failure. Circ Res 2013;113:646-59. [Crossref] [PubMed]
- Rosa-Garrido M, Chapski DJ, Schmitt AD, et al. High-Resolution Mapping of Chromatin Conformation in Cardiac Myocytes Reveals Structural Remodeling of the Epigenome in Heart Failure. Circulation 2017;136:1613-25. [Crossref] [PubMed]
- Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet 2016;17:661-78. [Crossref] [PubMed]
- Croft JA, Bridger JM, Boyle S, et al. Differences in the Localization and Morphology of Chromosomes in the Human Nucleus. J Cell Biol 1999;145:1119-31. [Crossref] [PubMed]
- Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 2015;16:245-57. [Crossref] [PubMed]
- Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet 2016;17:17-43. [Crossref] [PubMed]
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 2014;15:234-46. [Crossref] [PubMed]
- Mishra A, Hawkins RD. Three-dimensional genome architecture and emerging technologies: looping in disease. Genome Med 2017;9:87. [Crossref] [PubMed]
- Gomez-Velazquez M, Badia-Careaga C, Lechuga-Vieco AV, et al. CTCF counter-regulates cardiomyocyte development and maturation programs in the embryonic heart. PLoS Genet 2017;13:e1006985. [Crossref] [PubMed]
- Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev 2016;30:881-91. [Crossref] [PubMed]
- Monte E, Rosa-Garrido M, Karbassi E, et al. Reciprocal Regulation of the Cardiac Epigenome by Chromatin Structural Proteins Hmgb and Ctcf: IMPLICATIONS FOR TRANSCRIPTIONAL REGULATION. J Biol Chem 2016;291:15428-46. [Crossref] [PubMed]


