Atypical antibody responses to influenza
Introduction
Influenza virus is an RNA virus that circulates commonly among humans, causing acute respiratory infections. Each year it is estimated that three to five million people are infected with influenza virus, causing a substantial health burden and economic impact through loss of productivity (1). The young and the elderly are particularly susceptible to severe disease (2). Additionally, completely new antigenic strains can emerge from animal reservoirs through reassortment of the segmented viral genome to cause influenza pandemics (3). For these reasons, influenza viruses pose a constant and significant public health threat.
Antigenic classification of influenza viruses
Influenza viruses belong to the family Orthomyxoviridae and can be classified into four distinct types: influenza A, influenza B, influenza C and the newly identified (provisionally named) influenza D (4,5). Although Influenza A, B, and C viruses commonly circulate and cause disease in humans, only Influenza A and B are of significant concern [influenza C is usually only associated with mild respiratory infections in children (6)]. Due to their ability to rapidly evolve, influenza A and B viruses undergo antigenic drifts to cause annual seasonal epidemics. This, along with the specificity of the induced antibody response, necessitates annual influenza vaccination against these seasonal influenza viruses.
Influenza A virus (IAV) has been the cause of some of the most devastating infectious outbreaks in history (7). Aquatic birds are the natural reservoir of most, if not all, IAV and it is from this reservoir that viruses sporadically infect other hosts, sometimes establishing stable lineages within the new host species. IAV is classified into distinct subtypes based on its two major surface glycoproteins: hemagglutinin (HA) and neuraminidase (NA). The combination of HA and NA subtypes form the diverse strains of IAV and are the major antigenic targets of the host humoral immunity. HA is the most abundant protein on the virus surface and is responsible for binding the cellular receptor and mediating entry into the host cell (8). It is a tetrameric protein and each monomer contains a globular head and a stalk domain (9) (Figure 1A). Though the viral functionality of the HA protein is conserved, it can be phylogenetically and antigenically distinguished into multiple different subtypes. Currently, there are 18 subtypes of IAV HA (H1–H18) and 11 subtypes of IAV NA (N1–N11) identified, although the newest identified subtypes, H17, H18, N10 and N11 are of bat-origins (10). The IAV HA proteins are subclassified into two groups based on phylogenetic similarities: group 1 consists of H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17 and H18 while group 2 consists of H3, H4, H7, H10, H14 and H15 (Figure 1B).
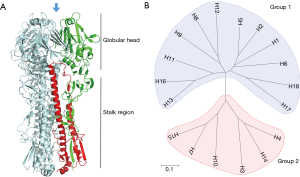
Aside from genetic and antigenic classification, the HA proteins can also be distinguished based on their receptor binding preference. Human strains of influenza virus recognizes the α(2,6)-linked sialic acid on host cells while avian strains preferentially binds the α(2,3)-linked sialic acid. Some swine-origin influenza viruses can also recognize both moieties (11,12).
The globular head of the HA molecule contains the receptor binding site where the virus attaches to sialic acid on the surface of cells to initiate infection. As the main mechanism of host invasion and the most protrusive molecule on the virus, the head is the most targeted region by the humoral immune response (13). As a result of this immune pressure, the globular head region has the highest mutation rates of all viral proteins, helping evade antibodies targeting it (antigenic drift) (14). Due to antigenic distinction across the various subtypes of HA, the serological response against one subtype typically does not confer reactivity against another (15). In contrast, the HA stalk region is more conserved and antibodies targeting this region are often capable of neutralizing influenza viruses from different IAV subtypes within the same phylogenetic group, and less commonly across groups 1 and 2 (Figure 1B) (16,17). Highlighting the conserved nature of stalk epitopes, one antibody, CR9114, has been identified that binds to the stalk region of influenza A and B (18). These cross-reactive antibodies, termed broadly neutralizing antibodies, are the subject of intense research as they represent a strategy to counter the threat of a diverse and highly mutable virus. For an up-to-date review of broadly neutralizing influenza antibodies and its mechanisms of action, see Corti et al. (17,19).
The B-cell responses to influenza virus exposure
Infection with any pathogen elicits an innate followed by an adaptive immune response. The adaptive immune response is mediated primarily by lymphocytes recognizing antigens, or more specifically, epitopes specific to the infecting pathogens. During a primary infection, where an antigen is encountered for the first time, an antibody response is the last line of immune defense to develop.
Antibodies are secreted by B-lymphocytes. B-cells are distinguishable from other lymphocytes by a B-cell receptor (BCR) on the cell surface, which is composed of an immunoglobulin (Ig) and an Ig-alpha/Ig-beta heterodimer. The Ig molecule contains a unique receptor to recognize a single cognate antigen, and is called an antibody when secreted (20,21).
During infection, an early T-helper cell-independent B-cell response generates short-lived effector B-cells that secrete low-affinity IgM or IgD antibodies. These antibodies provide early control of infection. A later response involves the engagement of T-helper cells to activate B-cells that lie within the germinal centers (GC) of the lymphoid tissues. This process, termed the GC reaction, causes the activated B-cell to undergo affinity maturation where they proliferate extensively while undergoing somatic hypermutation and class-switching to select for clones that bind the target antigen with high affinity. As these mature B cells proliferate, they are differentiated into distinct lineages of either long-lived, class switched effector “plasma” cells whose function is to secrete antibodies (of IgG, IgA or IgE isotype) or memory B-cells that are specific for the invading pathogen.
During primary infection, this B-cell response is achieved in about four weeks after initial infection. Upon resolution of infection, a period of cell death follows, after which only long-lived plasma and memory B-cells will remain (22,23). However, during secondary infection, the memory B-cells are activated rapidly to undergo clonal selection and affinity maturation, resulting in maximum antibody titer being secreted in a much shorter time compared to the primary infection (23). The increased accumulation of antibodies in the blood, termed seroconversion, can be detected via immunological assays and the rise in antibody titers is indicative of recent antigen exposure. The two most popular assays used for detecting seroconversion to influenza are the hemagglutination inhibition (HAI) assay, which allows detection of antibodies targeting the globular head of HA and the neutralization assay, which detects antibodies of any specificity that are able to neutralize the virus (24).
Atypical antibody responses to influenza virus exposure
A typical antibody response after exposure to influenza antigen is dominated by antibodies that target the globular head of the HA. These antibodies are capable of neutralizing only the immunizing antigen and other antigenically similar viruses, likely of the same HA subtype (homosubtypic seroconversion). However, in recent years, atypical antibody responses have been documented where more cross reactive profiles have been observed. In this review, we focus on the phenomena of atypical antibody responses after influenza exposures that result in seroconversion to a wider range of HAs. This includes induction of antibody responses to virus subtype that are different from the immunizing antigen (heterosubtypic seroconversion). The focus of this review is strictly on the serological response, and no other forms of heterotypic immunity such as that afforded by T-cells or other less antigenically-specific immune mechanisms. More specifically, we will consider heterosubtypic antibody responses as measured by a four-fold increase in antibody titer (seroconversion) to a non-infecting/exposed strain when measured by hemagglutination-inhibition assay (HAI) or neutralization assay.
Heterosubtypic antibody responses after infection
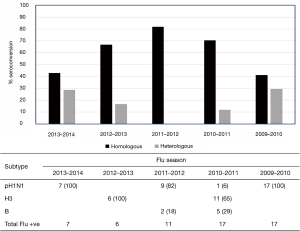
We became interested in this phenomenon after conducting a serological study on patients enrolled through a longitudinal-observational cohort study that was based at the Le Bonheur Hospital in Memphis, Tennessee (25). This cohort was initiated after the 2009 pandemic, and enrolled more than 300 participants over 5 influenza seasons. In this study, patients were tested for influenza positivity and subsequently subtyped by polymerase-chain-reaction (PCR). Serological testing was performed against H1N1, H3N2 and influenza B strains that were likely to be circulating in the Northern Hemisphere for that particular season. Our data revealed a small subset of individuals that showed an atypical antibody response (Figure 2) by seroconverting to more than one influenza subtype after infection. Although this heterosubtypic seroconversion first caught our attention after the 2009 pandemic, we noted that it occurred at varying degrees over the next 4 influenza seasons (Figure 2). A review of published literature revealed that this phenomenon had also been observed by others, albeit in very limited studies (26-28). The first of these studies were reported by Baz et al. (26) in a Canadian cohort during the 2009 H1N1 pandemic. They reported that 8 out of 67 (12%) individuals infected with the 2009 pandemic strain A(H1N1)pdm also seroconverted to the seasonal A/Brisbane/59/2007 (H1N1) strain, which was antigenically distinct from A(H1N1)pdm. More intriguingly however, 5 individuals that were positive by PCR and serology to A(H1N1)pdm also seroconverted to the seasonal H3N2 strain, A/Panama/2007/99. Similarly, a seroepidemiological study of antibody responses to A(H1N1)pdm in a Singaporean cohort identified 20%, 18% and 16% of the A(H1N1)pdm—confirmed cases (N=45) that seroconverted to antigenically-distinct A/Brisbane/59/2007 (H1N1), A/Brisbane/10/2007 (H3N2), and A/Wisconsin/15/2009 (H3N2), respectively (27). The authors also drew attention to a similar observation made in Singapore during the 1968 H3N2 pandemic, whereby at least 4 adults showed concomitant HAI-seroconversion to the newly emerged pandemic strain A/Hong Kong/1/1968 (H3N2) and the circulating A/Singapore/1/1957 (H2N2) strain (28).
Heterosubtypic antibody response after vaccination
Atypical antibody responses have also been reported in vaccine trials. For example, in the early vaccine trials of an A/Viet Nam/1203/2005 (H5N1)-based vaccine, 15 participants (3% of entire cohort) had positive H5 HAI-antibody titers at baseline (29). As the study was conducted in United States, it was highly unlikely that these individuals have been exposed to the H5N1 strain that was circulating only in Asia at that time.
How then do these individuals have H5N1 antibodies, particularly those that target the globular head, without prior exposure? Cross-reactive neutralizing antibodies have been shown to exist at low levels in pre-immune human serum (30) and can be boosted after vaccination with influenza strains possessing a divergent globular head (17,31-33). For example, during vaccine trials with an A(H1N1)pdm monovalent inactivated vaccine, some recipients were shown to seroconvert to the antigenically distinct seasonal A(H1N1) strains. However, those that were vaccinated with seasonal A(H1N1) did not have any serological response to the A(H1N1)pdm (34-36). Analysis of the post-vaccination activated B-cell clones suggested that vaccination of these individuals who had pre-existing immunity generated to antigenically distinct viruses preferentially selects for stalk-reactive memory B-cell clones, thus enhancing cross-reactive antibody titers (35). However, much of these stalk-reactive antibodies were only detectable by neutralization assays and not HAI assay. Furthermore, stalk antibodies typically do not occur at high levels within the host compared to antibodies that target the globular head (31). Thus, the HAI-positivity against H5N1 seen in the vaccine trial remains somewhat an enigma.
Co-infections or an immunological phenomenon?
The fact that all infection-associated heterosubtypic seroconversion events were reported during a pandemic raises an interesting question: are heterosubtypic seroconversions a consequence of dramatic antigenic shift or merely undetected incidences of co-infections? In the published studies, circulation of seasonal influenza strains was negligible, or absent altogether during the peak of the pandemic activity (Figure 3), suggesting that co-infection was unlikely. However, co-infection of a single individual with A(H1N1)pdm and seasonal influenza strains did occur during the 2009 pandemic (37,38). Historically, the most striking epidemiological support for co-infections was the emergence of the H1N2 virus that had limited global spread between 2000 and 2003 [reviewed in (39)]. This reassortant virus was generated from an A/Moscow/10/1999 (H3N2)-like virus that had acquired the HA from an A/New Caledonia/20/1999 (H1N1)-like virus (40).
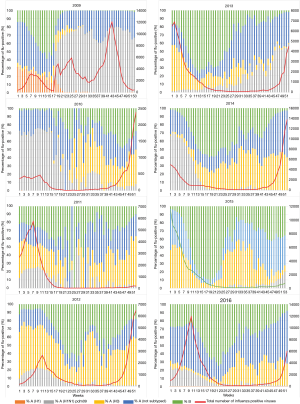
Analysis of influenza subtype circulation in the United States between 2009 and 2016, suggests that a single subtype typically dominates at greater than 80% prevalence during peak activity in a typical influenza (non-pandemic) season. The exception to this was in the year after the pandemic (the 2010–2011 influenza season) where both the A(H1N1)pdm and H3N2 subtypes were detected at almost similar proportions (Figure 3). Thus, epidemiological data at the population level suggests that co-circulation of multiple subtypes certainly can occur during some influenza seasons.
How often then, are co-infections within an individual detected? Review of the few published manuscripts suggests that co-infections by different influenza subtypes are only rarely detected; studies by Perez-Garcia and Falchi reported a 1.6%- and 3.2% co-infection rates respectively (41,42). The highest co-infection rate (7.3%) was reported by Goka et al., from over 25,000 respiratory samples analyzed over a period of 4 years (43). While these data suggest that co-infection can be a plausible explanation for heterosubtypic seroconversion, the lack of a reliable estimate of its occurrence and any systematic study to link this precludes any conclusive association. Furthermore, co-infection will not explain the observation in the study by Yin-Murphy (28), whereby seroconversion was detected against a strain that was no longer in circulation.
Original antigenic sin
The possibility that these heterosubtypic seroconversions may be a result of an immunological phenomenon should be considered. It is now evident that the antibody response to influenza is complex due to the influence of the host memory response and the antigenic variability in the HA. The ability to produce HA stalk-reactive antibodies in the absence of a highly similar globular head has been attributed to the effects of original antigenic sin (OAS) (17). OAS, first coined by Thomas Francis (44), refers to the phenomenon in which, following exposure via vaccination or infection to a virus that is antigenically similar to a previously encountered strain, the body will preferentially recall the originally encountered memory B-cell clones, resulting in an increase in antibody response to the original antigen.
Two recent studies have provided intriguing new insights into the effects of OAS after influenza vaccination. In the first, Huang et al. showed that although the induced antibody response upon influenza vaccination is polyclonal, a majority of these antibody clones recognized epitopes that were common to the strain that the host was most likely to have been first exposed to (45). The authors proposed that OAS was the reason why the donor made antibodies that failed to neutralize recently emerged strains that possess a single major mutation that had evaded neutralization. In a similar vein, the second study by Schmidt et al. showed the germline B-cell precursor, the “unmutated common ancestors” (UCA) of six clonal lineage of broadly neutralizing antibodies produced antibodies that recognized strains that the host would most likely have been exposed to. Upon vaccination however, the increased breadth of reactivity was due to clonal proliferation and diversification of the original clone (46). These two studies illustrate how the memory B-cell response can impact the diversity and breadth of antibody response upon re-exposure to influenza virus antigen.
However, while OAS effects can result in a misdirected antibody response during infection or vaccination, it cannot yet account for the heterosubtypic seroconversion against multiple antigenically distinct subtypes observed after infection.
Broad B-cell repertoire or polyreactive antibodies?
That certain individuals can produce significant titers against cross-reactive epitopes suggests that these individuals, for some unknown reason, may have a broader B-cell repertoire than normal. Intriguingly, researches in the broadly neutralizing antibodies field have shown that the ability to mount these broadly neutralizing antibodies was also linked to a host-genetic component. For example, generation of the Group 1-broadly neutralizing antibodies often involve somatic hypermutation and usage of the Ig heavy-chain variable region VH1-69 gene (32,47). Indeed, a germline encoded polymorphism in this gene was an early requirement for the generation of these antibodies (48). Thus it may be possible that a host-genetic component could predispose certain individuals to generate a highly diverse B-cell repertoire after infection.
Another possible immunological phenomenon that could underlie the atypical antibody response is the generation of polyreactive antibodies. Polyreactive antibodies or “natural antibodies” as its name implies, exist as part of the normal immune repertoire. They can bind to multiple ligands without needing prior antigen exposure. Due to their lack of ligand-specificity, they are considered to have “innate” immune-like function. Although polyreactive antibodies are typically of low affinity and of the IgM-isotype, high affinity polyreactive IgG and IgA isotypes have also been described [reviewed in (49)]. It is important to recognize that these antibodies do not typically exist in high titers as a safeguard against self-reactivity. Polyreactive B-cell clones are generally selected against during the B-cell maturation process, except in malignancies in which this process becomes impaired (i.e., such as in systemic lupus erythematosus, SLE).
Polyreactive antibodies have been described after bacterial and viral infections; and have particularly well-studied in the context of human immunodeficiency virus (HIV). It was found that some broadly neutralizing HIV-antibodies are also polyreactive against self-antigens (50,51). One salient feature of these antibodies is the high-plasticity of their antigen-binding pocket that results in more permissive binding of different ligand structures (such as was described for the influenza A and B cross-reactive antibody CR9114). Despite the wealth of literature for HIV, no similar observations have been made for influenza. The only exception was a study by Kaur et al., which found no statistically significant differences in the levels of broadly neutralizing or polyreactive influenza antibodies in a cohort of influenza vaccinated SLE patients compared to controls, although there were certainly intriguing trends detected (52).
Current knowledge gaps and future research directions
While there have been insightful studies into the mechanisms and requirements that drive broadly influenza virus-neutralizing antibody responses—particularly after influenza vaccinations—it is unknown if the same processes underlie heterosubtypic antibody responses seen after naturally acquired influenza virus infections. Furthermore, it is currently unclear whether individuals with heterotypic responses are better protected against infection by diverse influenza virus subtypes.
The studies cited in this review all reported heterosubtypic seroconversion events during a pandemic. Whether this was a chance observation due to increased surveillance and serological testing or a pandemic-associated phenomenon is unknown. Hence, a critical knowledge gap currently is in determining the prevalence of heterosubtypic seroconversion during a typical influenza season. This type of study presents obvious logistical and economical challenges. At the reported prevalence rate of co-infections, any studies attempting to examine this phenomenon will need to enroll a large cohort during an influenza season. Extensive molecular detection and serological analysis against diverse strains will need to be performed to detect these heterosubtypic seroconversion events. When a baseline prevalence rate is established, other factors such as age, subtype, and underlying conditions can be examined to determine if this is an immunological or virological phenomenon.
Summary
Atypical antibody responses have been reported in the context of influenza virus infection and vaccination. Many of these reports have focused on the effects that the 2009 A(H1N1)pdm had on eliciting heterotypic antibody response. These studies suggest that the 2009 pandemic virus or exposure to antigenically shifted virus with no prior immunity may be apposite for inducing heterosubtypic seroconversions. However, there have also been vaccination studies in which the influenza vaccine was able to generate a heterosubtypic response. While some of these observations have an immunological basis, others such as heterosubtypic seroconversion events observed after infection still lack a satisfying explanation. While it is possible that OAS, genetic predisposition to form broad B-cell repertoires, polyreactive antibodies, and/or other currently undescribed immune mechanisms may play a role in these responses, we are still lacking direct evidence to suggest a mechanism. Heterosubtypic seroconversion hitherto represents an unknown and largely unstudied phenomenon of the immune response that should be explored further. If indeed such responses are able to create protection from unencountered strains, the mechanism by which these individuals are able to significantly broaden their antibody repertoire may prove useful both for general vaccine design and pandemic prevention.
Acknowledgements
We would like to thank Dr. Robert Webster and Dr. Paul Thomas from St. Jude Children’s Research Hospital for critical reading of the manuscript. We would like to thank Dr. Mark Zanin for his help in generating the figures.
Funding: This work is supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (contract no. HHSN266200700005C) and by the American Lebanese Syrian Associated Charities.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 2007;25:5086-96. [Crossref] [PubMed]
- Organization WH. Influenza (Seasonal) Fact Sheet. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/
- Guan Y, Vijaykrishna D, Bahl J, et al. The emergence of pandemic influenza viruses. Protein Cell 2010;1:9-13. [Crossref] [PubMed]
- Andrewes CH, Bang FB, Burnet FM. A short description of the Myxovirus group (influenza and related viruses). Virology 1955;1:176-84. [Crossref] [PubMed]
- Hause BM, Collin EA, Liu R, et al. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio 2014;5:e00031-14. [Crossref] [PubMed]
- Matsuzaki Y, Katsushima N, Nagai Y, et al. Clinical features of influenza C virus infection in children. J Infect Dis 2006;193:1229-35. [Crossref] [PubMed]
- Webby RJ, Webster RG. Emergence of influenza A viruses. Philos Trans R Soc Lond B Biol Sci 2001;356:1817-28. [Crossref] [PubMed]
- Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem 1987;56:365-94. [Crossref] [PubMed]
- Green N, Alexander H, Olson A, et al. Immunogenic structure of the influenza virus hemagglutinin. Cell 1982;28:477-87. [Crossref] [PubMed]
- Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog 2013;9. [Crossref] [PubMed]
- Connor RJ, Kawaoka Y, Webster RG, et al. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 1994;205:17-23. [Crossref] [PubMed]
- Gambaryan AS, Tuzikov AB, Piskarev VE, et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6'-sialyl(N-acetyllactosamine). Virology 1997;232:345-50. [Crossref] [PubMed]
- Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology 2006;119:1-7. [Crossref] [PubMed]
- Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine 2007;25:6852-62. [Crossref] [PubMed]
- Couch RB, Kasel JA. Immunity to influenza in man. Annu Rev Microbiol 1983;37:529-49. [Crossref] [PubMed]
- Krammer F, Hai R, Yondola M, et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol 2014;88:3432-42. [Crossref] [PubMed]
- Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011;333:850-6. [Crossref] [PubMed]
- Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science 2012;337:1343-8. [Crossref] [PubMed]
- Corti D, Cameroni E, Guarino B, et al. Tackling influenza with broadly neutralizing antibodies. Curr Opin Virol 2017;24:60-9. [Crossref] [PubMed]
- Parker DC. T cell-dependent B cell activation. Annu Rev Immunol 1993;11:331-60. [Crossref] [PubMed]
- Reth M. The B-cell antigen receptor complex and co-receptors. Immunol Today 1995;16:310-3. [Crossref] [PubMed]
- Ennis FA, Rook AH, Qi YH, et al. HLA restricted virus-specific cytotoxic T-lymphocyte responses to live and inactivated influenza vaccines. Lancet 1981;2:887-91. [Crossref] [PubMed]
- Murphy BR, Nelson DL, Wright PF, et al. Secretory and systemic immunological response in children infected with live attenuated influenza A virus vaccines. Infect Immun 1982;36:1102-8. [PubMed]
- Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther 2011;9:669-83. [Crossref] [PubMed]
- Oshansky CM, Gartland AJ, Wong SS, et al. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am J Respir Crit Care Med 2014;189:449-62. [Crossref] [PubMed]
- Baz M, Papenburg J, Hamelin ME, et al. Seroconversion to seasonal influenza viruses after A(H1N1)pdm09 virus infection, Quebec, Canada. Emerg Infect Dis 2012;18:1132-4. [Crossref] [PubMed]
- Chen MI, Barr IG, Koh GC, et al. Serological response in RT-PCR confirmed H1N1-2009 influenza a by hemagglutination inhibition and virus neutralization assays: an observational study. PLoS One 2010;5. [Crossref] [PubMed]
- Yin-Murphy M. An outbreak of "hong Kong 'flu" in Singapore. II. Virological and serological report. Singapore Med J 1970;11:33-7. [PubMed]
- Treanor JJ, Campbell JD, Zangwill KM, et al. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N Engl J Med 2006;354:1343-51. [Crossref] [PubMed]
- Sui J, Sheehan J, Hwang WC, et al. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis 2011;52:1003-9. [Crossref] [PubMed]
- Ellebedy AH, Krammer F, Li GM, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A 2014;111:13133-8. [Crossref] [PubMed]
- Joyce MG, Wheatley AK, Thomas PV, et al. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 2016;166:609-23. [Crossref] [PubMed]
- Whittle JR, Wheatley AK, Wu L, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 2014;88:4047-57. [Crossref] [PubMed]
- Cortina-Ceballos B, Godoy-Lozano EE, Tellez-Sosa J, et al. Longitudinal analysis of the peripheral B cell repertoire reveals unique effects of immunization with a new influenza virus strain. Genome Med 2015;7:124. [Crossref] [PubMed]
- Li GM, Chiu C, Wrammert J, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 2012;109:9047-52. [Crossref] [PubMed]
- Thomson CA, Wang Y, Jackson LM, et al. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Front Immunol 2012;3:87. [Crossref] [PubMed]
- Peacey M, Hall RJ, Sonnberg S, et al. Pandemic (H1N1) 2009 and seasonal influenza A (H1N1) co-infection, New Zealand, 2009. Emerg Infect Dis 2010;16:1618-20. [Crossref] [PubMed]
- Rith S, Chin S, Sar B, et al. Natural co-infection of influenza A/H3N2 and A/H1N1pdm09 viruses resulting in a reassortant A/H3N2 virus. J Clin Virol 2015;73:108-11. [Crossref] [PubMed]
- Komadina N, McVernon J, Hall R, et al. A historical perspective of influenza A(H1N2) virus. Emerg Infect Dis 2014;20:6-12. [Crossref] [PubMed]
- Xu X, Smith CB, Mungall BA, et al. Intercontinental circulation of human influenza A(H1N2) reassortant viruses during the 2001-2002 influenza season. J Infect Dis 2002;186:1490-3. [Crossref] [PubMed]
- Falchi A, Arena C, Andreoletti L, et al. Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. J Clin Virol 2008;41:148-51. [Crossref] [PubMed]
- Perez-Garcia F, Vasquez V, de Egea V, et al. Influenza A and B co-infection: a case-control study and review of the literature. Eur J Clin Microbiol Infect Dis 2016;35:941-6. [Crossref] [PubMed]
- Goka E, Vallely P, Mutton K, et al. Influenza A viruses dual and multiple infections with other respiratory viruses and risk of hospitalisation and mortality. Influenza Other Respir Viruses 2013;7:1079-87. [Crossref] [PubMed]
- Francis TJ. On the Doctrine of Original Antigenic Sin. Proc Am Philos Soc 1960;104:572.
- Huang KY, Rijal P, Schimanski L, et al. Focused antibody response to influenza linked to antigenic drift. J Clin Invest 2015;125:2631-45. [Crossref] [PubMed]
- Schmidt AG, Do KT, McCarthy KR, et al. Immunogenic Stimulus for Germline Precursors of Antibodies that Engage the Influenza Hemagglutinin Receptor-Binding Site. Cell Rep 2015;13:2842-50. [Crossref] [PubMed]
- Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009;16:265-73. [Crossref] [PubMed]
- Pappas L, Foglierini M, Piccoli L, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014;516:418-22. [Crossref] [PubMed]
- Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci 2012;69:1435-45. [Crossref] [PubMed]
- Haynes BF, Fleming J, St Clair EW, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 2005;308:1906-8. [Crossref] [PubMed]
- Mouquet H, Scheid JF, Zoller MJ, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 2010;467:591-5. [Crossref] [PubMed]
- Kaur K, Zheng NY, Smith K, et al. High Affinity Antibodies against Influenza Characterize the Plasmablast Response in SLE Patients After Vaccination. PLoS One 2015;10. [Crossref] [PubMed]


