Bronchoalveolar lavage fluid microbiota dysbiosis in infants with protracted bacterial bronchitis
Introduction
Protracted bacterial bronchitis (PBB), which features a wet cough, persistent infection of the bronchial lining and mucopurulent inflammation, is one of the main causes of recurrent coughing and wheezing in infants and toddlers (1,2). PBB is considered as forerunner of other chronic respiratory diseases like bronchiectasis and chronic suppurative pneumonia (3,4). Bacterial culture has shown Haemophilus influenza, Streptococcus pneumonia and Moraxella catarrhalis to be the main pathogens (1,2). However, the low detection rate of the traditional method limits the discovery of pathogens present at low abundance. Assisted by next generation of sequencing, 16S rDNA detection of microbiota provides more complete information regarding the flora of the respiratory tract in PBB infants (3-5).
Bronchoalveolar lavage fluid (BALF) is normally used to diagnose and characterize diseases of the lower respiratory tract (6-8). Using 16S rDNA detection on BALF, the bacterial constitution and microbiota diversity in the lower respiratory tracts of infants with PBB can be described precisely (7-9). In a study reported by van der Gast et al., the detection on BALF microbiota indicated that PBB infants shared common core bacteria with infants with bronchiectasis and infants with cystic fibrosis (10), and the infected pathogens would damage the bronchial mucosa continuously. Whilst Wang et al. suggested that the composition of BALF microbiota differed in infants with pneumonia infected with different pathogens, and the interactions among bacteria were described in detail (11). There have been few reports of BALF microbial comparison between infants with PBB and tracheomalacia (TM), and the impact of co-occurrence network on infants with PBB has not yet been assessed.
In this study, BALF, which was collected from 24 infants, was used for microbiota detection and co-occurrence network construction in PBB and TM infants. And two issues need to be resolved: (I) bacterial composition differences between PBB and TM infants; (II) microbial interactions in PBB infants and their contributions to overall health. This would deepen our understanding of the bacterial network and its involvement in the pathogenesis of PBB.
Methods
Ethics statement and clinical diagnosis
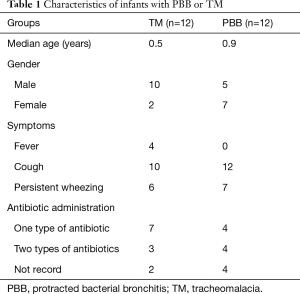
All infants’ parents provided written informed consent, and the study was approved by the Ethical Committee of Shenzhen Children’s Hospital under approval number 2016(005). PBB was defined as persistent bacterial infection of the bronchial epithelium, which induces such clinical features as chronic purulent inflammation. The PBB infants should meet the following conditions: (I) the patients should have had a chronic wet cough lasting at least 4 weeks; (II) the cough symptoms should be relieved after 2 weeks of antibiotic treatment; (III) there were more neutrophils in patients with positive bacterial cultures; (IV) other causes of chronic cough were excluded (12). The TM infants who fulfilled the following criteria were included in this study: (I) the patients have more than 50% tracheal lumen collapse; (II) clinical symptoms, which including noisy respiration, tracheal rhonchi, harsh barking cough or expiratory dyspnea, can be observed; (III) the problems of cardiac diseases, neurological disorders and esophageal abnormalities should be ruled out (13). All patients’ clinical presentations and medication histories were summarized in Table 1 and Table S1.
Full table

Full table
BALF collection
After admission to our hospital, BALF was collected from the infants in 2 days. An electronic bronchoscope (EB-270P or EB-270S, Fujitsu Electronics, Inc., Tokyo, Japan) was used for sampling. The infants were forbidden to eat 6 h before operation. Atropine (0.01–0.02 mg/kg) was injected into the patients intramuscularly 30 min before operation, and then midazolam (0.1–0.3 mg/kg) was applied intravenously. After the induction of electronic bronchoscopes, lidocaine (1–2 mL) with concentration of 1% was sprayed on the main bronchi. Then the electronic bronchoscopes were fixed at middle lobe, lower bronchus of upper lobe and inflamed parts of the patients’ right lungs. Saline (1 mL/kg) was used for bronchoalveolar lavage. The BALF was reclaimed and stored at −80 °C for further use.
Library construction and DNA sequencing
Microbial DNA was extracted from BALF using the E.Z.N.A.® Soil DNA Kit (Omega Bio-tek, Inc., Norcross, GA, USA) according to manufacturer’s protocols. Using a PCR kit (AP221-02, Beijing TransGen Biotech Co., Ltd., Beijing, China), 16S rDNA V3–V4 regions of samples were amplified using primers 341F and 517R. Agarose gel electrophoresis with 2% concentration was used to confirm the length of polymerase chain reaction (PCR) products, and the quality of PCR products was detected by QuantiFluor™-ST [Promega (Beijing) Biotech Co., Ltd., Beijing, China]. Then the qualified DNA was used for MiSeq sequencing with paired-end 300 strategies. Raw reads were uploaded to NCBI Sequence Read Archive (SRA) Database (accession number: SRP067201, SRR3951741, SRR3951743, and SRR3951754).
Data processing and taxonomical annotation
Raw data, which were obtained by MiSeq sequencing, was used for data filtering firstly. Any reads that contained more than 10 low-quality bases or 15 bp adapter sequences were filtered out. Duplications were also removed to leave behind clean, high-quality data. Then the paired reads were connected into tags based on overlapping. The tags were clustered into operational taxonomic units (OTUs) with 97% similarity as indicated by USEARCH (v7.0.1090) (14) and each OTU contained one raw representative sequence (15). Chimeras, which created during PCR, were removed from OTUs using UCHIIME (16). Finally, all the tags were mapped to the OTUs using search global and 466 final OTUs were obtained in total online: http://jtd.amegroups.com/public/addition/jtd/supp-jtd.2017.12.59-2.pdf (15). The Shannon and Simpson value was calculated using MOTHUR (v1.31.2) (17).
Statistical analysis
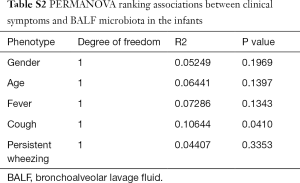
The diversity of respiratory tract microbiota between PBB and TM group was compared by using Wilcoxon rank-sum test. The significantly enriched microbial residents in PBB and TM group were identified through LEfSe [linear discriminant analysis (LDA) effect size] analysis, according to the following parameters: the P values of Kruskal-Wallis test and Wilcoxon test were smaller than 0.01 and the cutoff of LDA score was set as 3 (18). Using package “psych” in R, Pearson correlations among genera based on their relative abundances were obtained in PBB and TM cohorts. Then the co-abundance network was visualized with Cytoscape software (v2.2.0) (19) if the Pearson correlation coefficients >0.9 or <−0.5. Using Bray-Curtis distances between PBB and TM infants, the effect of clinical symptoms on BALF microbial composition were detected using permutational analysis of variance (PERMANOVA, 9,999 permutations) with package “vegan” in R (Table S2).
Full table
Results
Study cohorts and data output
A total of 12 infants with PBB and 12 infants with TM, who were younger than 3 years old, were enrolled for BALF microbiota detection. And their clinical information was exhibited in Table 1. Using MiSeq platform targeting of 16S rDNA V3–V4 variable region, the tags of BALF samples were obtained and ranged from 9,992 to 29,165. The number of OTUs ranged from 75 to 117 in PBB group and 79 to 245 in TM group.
Microbiota diversity of BALF was significantly lower in PBB cohort
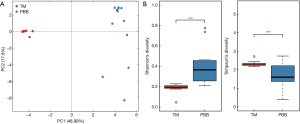
Principal component analysis (PCA) based on OTU distribution was performed to assess the microbiota similarity among 24 samples online: http://jtd.amegroups.com/public/addition/jtd/supp-jtd.2017.12.59-4.pdf. Results showed that samples from PBB group clustered together, as did those phenomenons from TM group (Figure 1A). Then Shannon and Simpson indexes were used to evaluate the BALF microbial diversity. The average value of Shannon index was 1.683±0.703 (mean ± SD) for PBB group and 2.324±0.142 for TM group (P<0.001) (Figure 1B). The significant difference was also indicated by Simpson index, which averaged 0.416±0.216 for PBB group and 0.191±0.025 for TM group (P<0.001) (Figure 1B).
Taxonomic components differed between PBB and TM infants
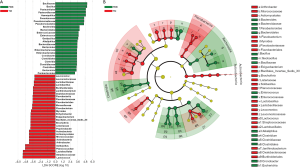
Using LEfSe analysis, the composition of BALF microbiota was compared and 54 differentially enriched taxa were discovered between PBB and TM group (Figure S1A). Among the 25 taxa enriched in the PBB group, Bacillus (40.770%±22.218%, LDA =5.632), Haemophilus (14.319%±29.532%, LDA =5.161), Pseudomonas (10.406%±25.439%, LDA =5.042) and Enterococcus (0.959%±0.631%, LDA =3.980) were present at higher relative abundances. The relative abundance of Lactobacillales, which includes Lactococcus (13.463%±7.319%, LDA =5.661) and Lactobacillus (0.153%±0.076%, LDA =3.508), was significantly lower in PBB infants. To further characterize the taxonomic differences, the BALF microbiota was analyzed at the class level. Consistent with results for genera, the taxa enriched in PBB cohort were clustered on Bacteroidia and Clostridia, while Actinobacteria and Flavobacteria were more highly enriched in TM group (Figure S1B). The effect of potential confounders, including gender, age and clinical symptoms, on the alterations of BALF microbiota in PBB infants was evaluated with PERMANOVA (Table S2). And the correlation between cough and microbiota changes was identified (P<0.05).
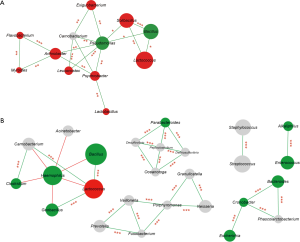
Distinctive BALF microbial co-occurrence network in PBB and TM children
For TM infants, Pseudomonas and Arthrobacter were the core nodes of the co-occurrence network and positively correlated with other genera (Figure 2). Meanwhile, it has been suggested that significantly enriched Lactococcus was positively correlated with Bacillus (r=0.920, P<0.001). In PBB group, the aforementioned network was destroyed and a more complicate microbiota co-occurrence network was exhibited (Figure 2). Four core nodes were shown, including Haemophilus, Peptoclostridium, Porphyromonas and Cronobacter. Haemophilus, which was the main causative pathogen of PBB, was negatively correlated with other genera, including Bacillus (r=−0.533), Clostridium (r=−0.541) and Lactococcus (r=−0.539). The positive correlation between Lactococcus and Bacillus (r=0.940, P<0.001) in PBB group tended to be similar to those in TM group.
Discussion
In the study, the composition of BALF microbial communities in PBB and TM patients was compared. PCA results have shown that the microbiota composition in PBB infants differed from that of TM infants. In PBB infants, the lung was invaded by pathogens, which might destruct the intact of respiratory mucosa and aggravate microbiota dysbiosis (20). Moreover, the correlation between clinical symptoms and microbial community changes proved that cough which caused by pathogen infection can be valued as the feature of PBB patients. And the infants with PBB had visibly lower microbial diversity. All the patients in the study were exposed to antibiotic treatments in the last 3 months, and lower microbial diversity in PBB infants was probably related with long-term antibiotic application although some clinical records cannot be accessed (Table S1).
Besides pathogen Haemophilus, the relative abundances of Pseudomonas, Escherichia, and Bacteroides were higher in PBB infants. These bacteria are difficult to culture especially after antibiotic treatment (20). Previous reports have demonstrated that Haemophilus was closely correlated with interleukin-1α (IL-1α) andinterleukin-1β (IL-1β) (21). They could trigger cell death and inflammation, which was responsible for the wet cough in PBB infants (12,21). Whilst, some studies suggested that Bacteroides could promote the differentiation of T helper cell 17 (Th17), and Th17 secreted interleukin-17 (IL-17) could trigger the inflammatory reaction and autoimmune diseases (22,23). As with PBB, there was evidence that the colonization of pathogenic bacteria such as Haemophilus in the airways was associated with severe airway obstruction and neutrophilic airway inflammation in adults with asthma (24). Pediatric PBB and adult asthma, which shared some common clinical manifestations and pathogens, gave us the idea that similar pathogenesis might existed in these two conditions. On the other hand, the abundance of Lactococcus and Lactobacillus was significantly lower in PBB infants. Lactococcus could secret butyrate and repress the growth of pathogens (25), while Lactobacillus was found to manipulate the development of lower airway flora in infants (26). In this way, the disorder of the lung microbiota in PBB infants can not only harm pediatric health directly but could also affect the colonization of bacteria in the lower airway and increase the risk of allergic airway disease.
The overall microbial co-occurrence network was destructed in PBB infants but not in TM cohorts. Significant enrichment of the core node Haemophilus was observed in infants with PBB, and it was negatively correlated with other microbial colonizers, including Lactococcus, Bacillus and Clostridium. Previous reports have shown that Bacillus affect the secretion of IL-17 indirectly (27), and Clostridium could promote T regulatory (Treg) cell differentiation, which were crucial for inflammation reactions (22,28). In infants with PBB, the relative abundances of aforementioned genera decreased as Haemophilus increased, and this was found to aggravate microbial imbalance and expand the immune response in the respiratory mucosa.
This study improved the understanding of differences in microbiota and bacterial co-occurrence networks between PBB and TM infants, but the lack of strong evidences for the relationship between lung microbiota and PBB pathogenesis was the major flaw of the study. Additional work is imperative: (I) large-cohort study should be performed to verify these findings; (II) microbiome and metabolite alterations need to be detected in PBB infants; (III) immune factor changes must be tested experimentally.
Conclusions
In summary, the study described the microbiota changes in lower respiratory tracts of PBB infants. The results have shed some preliminary lights on the manner in which the causative pathogens of PBB react, and it provides a foundation for bacterial adjunctive therapy of infantile PBB in accordance with antibiotic treatment.
Acknowledgements
We would like to thank the staff of WeHealthGene, whose names are not included in the list of authors but nonetheless contributed to the work of this team. We would also like to thank the nurses who helped collecting samples at the Shenzhen Children’s Hospital.
Funding: This work is funded by the Key Medical Disciplines Building Project of Shenzhen which was granted by Shenzhen Health and Family Planning Commission (201606033).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All infants’ parents provided written informed consent, and this study was approved by the Ethical Committee of Shenzhen Children’s Hospital under approval number 2016(005).
References
- Wang Y, Hao C, Chi F, et al. Clinical characteristics of protracted bacterial bronchitis in Chinese infants. Sci Rep 2015;5:13731. [Crossref] [PubMed]
- Chipps BE. Evaluation of infants and children with refractory lower respiratory tract symptoms. Ann Allergy Asthma Immunol 2010;104:279-83; quiz 283-5, 298.
- de Steenhuijsen Piters WA, Sanders EA, Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci 2015;370:20140294. [Crossref] [PubMed]
- Durack J, Boushey HA, Lynch SV. Airway Microbiota and the Implications of Dysbiosis in Asthma. Curr Allergy Asthma Rep 2016;16:52. [Crossref] [PubMed]
- Brown PS, Pope CE, Marsh RL, et al. Directly sampling the lung of a young child with cystic fibrosis reveals diverse microbiota. Ann Am Thorac Soc 2014;11:1049-55. [Crossref] [PubMed]
- Buzan MT, Pop CM. State of the art in the diagnosis and management of interstitial lung disease. Clujul Med 2015;88:116-23. [Crossref] [PubMed]
- Huffnagle GB, Dickson RP. The bacterial microbiota in inflammatory lung diseases. Clin Immunol 2015;159:177-82. [Crossref] [PubMed]
- Chen AC, Martin ML, Lourie R, et al. Adult non-cystic fibrosis bronchiectasis is characterised by airway luminal Th17 pathway activation. PLoS One 2015;10:e0119325. [Crossref] [PubMed]
- Yang XJ, Wang YB, Zhou ZW, et al. High-throughput sequencing of 16S rDNA amplicons characterizes bacterial composition in bronchoalveolar lavage fluid in patients with ventilator-associated pneumonia. Drug Des Devel Ther 2015;9:4883-96. [PubMed]
- van der Gast CJ, Cuthbertson L, Rogers GB, et al. Three clinically distinct chronic pediatric airway infections share a common core microbiota. Ann Am Thorac Soc 2014;11:1039-48. [Crossref] [PubMed]
- Wang H, Dai W, Qiu C, et al. Mycoplasma pneumoniae and Streptococcus pneumoniae caused different microbial structure and correlation network in lung microbiota. J Thorac Dis 2016;8:1316-22. [Crossref] [PubMed]
- Baines KJ, Upham JW, Yerkovich ST, et al. Mediators of neutrophil function in children with protracted bacterial bronchitis. Chest 2014;146:1013-20. [Crossref] [PubMed]
- Fraga JC, Jennings RW, Kim PC. Pediatric tracheomalacia. Semin Pediatr Surg 2016;25:156-64. [Crossref] [PubMed]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996-8. [Crossref] [PubMed]
- Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261-7. [Crossref] [PubMed]
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. [Crossref] [PubMed]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537-41. [Crossref] [PubMed]
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc 2014;11 Suppl 1:S21-7. [Crossref] [PubMed]
- Martin SJ. Cell death and inflammation: the case for IL-1 family cytokines as the canonical DAMPs of the immune system. FEBS J 2016;283:2599-615. [Crossref] [PubMed]
- Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013;13:321-35. [Crossref] [PubMed]
- Yurkovetskiy LA, Pickard JM, Chervonsky AV. Microbiota and autoimmunity: exploring new avenues. Cell Host Microbe 2015;17:548-52. [Crossref] [PubMed]
- Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One 2014;9:e100645. [Crossref] [PubMed]
- Ramakrishna BS. Role of the gut microbiota in human nutrition and metabolism. J Gastroenterol Hepatol 2013;28 Suppl 4:9-17. [Crossref] [PubMed]
- Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet 2013;381:861-73. [Crossref] [PubMed]
- Gong Y, Li H, Li Y. Effects of Bacillus subtilis on Epithelial Tight Junctions of Mice with Inflammatory Bowel Disease. J Interferon Cytokine Res 2016;36:75-85. [Crossref] [PubMed]
- Ireland SJ, Monson NL, Davis LS. Seeking balance: Potentiation and inhibition of multiple sclerosis autoimmune responses by IL-6 and IL-10. Cytokine 2015;73:236-44. [Crossref] [PubMed]


