COP9 signalosome subunit CSN5, but not CSN6, is upregulated in lung adenocarcinoma and predicts poor prognosis
Introduction
The COP9 signalosome (CSN) is an evolutionarily conserved complex which has been implicated in protein degradation, DNA damage response and signal transduction. This complex is composed of eight subunits designated as CSN1-CSN8. Among the eight subunits, CSN5 and CSN6 are the only two that each contain MPN (Mpr1p and Pad1p N-terminal) domain, the other six subunits CSN1, 2, 3, 4, 7, 8 contain PCI domain. The CSN exerts biological functions mainly through binding and inactivating the Cullin-RING E3 ubiquitin ligases (CRLs) which comprise over 250 members in human. The CSN is a Zinc2+-dependent isopeptidase which removes the ubiquitin-like protein, NEDD8, from Cullin in the CRLs. Structural analysis of CSN by X-ray crystallography has shown that with assistance of CSN4, CSN5-CSN6 heterodimer moves towards neddlyated CRLs and exerts isopeptidase activity (1). This complex can also bind the deneddylated CRLs and maintain them in an inactive state.
As the only two MPN domain-containing subunits, CSN5 and CSN6 share sequence and structural similarity. However, these two subunits also exhibit distinct features. The catalytic site and functional JAMM (JAB1/MPN/Mov34 metalloenzyme) motif are only present in CSN5, whereas CSN6 does not contain JAMM motif in its MPN domain. Another important feature is that CSN5 exists as two discrete forms in the cell: a holo-complex associated-form and monomeric form (the free form) (2), However, in substrate-free holo-complex, CSN5 is autoinhibited. Only when neddylated CRLs is binding to CSN and sensed by CSN4, CSN5 with the assistance of CSN6, can become functionally active (1,3). In contrast, CSN6 is tightly integrated into the CSN. The MPN domain of CSN6 and CSN5 form an intimate dimmer and the MPN domain of CSN6 is required in the stabilizing the structure of CSN5 MPN domain (4,5). CSN6 also interacts with every other subunit at the center of helical bundle which are formed by the carboxyl-terminal α-helices from all the subunits (1). The removal of CSN6 would result in the loss of structural integrity of the CSN (6), thus it is unlikely for CSN6 to release from the holo-complex (1).
Importantly, besides isopeptidase activity, emerging evidence demonstrates that CSN5 also possesses deubiquitination activity. Deubiquitination of HSP70 by CSN5 were shown to regulate exosomal protein sorting (7), and CSN5 was also reported to modulate the deubiquitination and stabilization of PD-L1 in response to inflammatory cytokines, thus control T cell suppression in cancer (8).
Recent studies demonstrated that CSN5 and CSN6 have been implicated in cancer development and progression. It has been reported that CSN5 and CSN6 were overexpressed in a variety of human cancers (9,10), such as lung cancer, breast cancer, ovarian cancer, thyroid cancer, colorectal cancer, hepatocellular carcinoma and glioblastoma (11,12). Their expressions correlate with clinical outcomes of the patients with cancer (13-15). Cellular and molecular mechanistic studies reveal that CSN5 and CSN6 are involved in ubiquitin-mediated degradation of important mediators implicated in cell cycle progression, signal transduction and apoptosis. For instance, both CSN5 and CSN6 was identified as a c-Jun activator (16,17). CSN5 and CSN6 are capable of inducing degradation of cyclin-dependent kinase inhibitor p27Kip1 and p57Kip2 (14,15,18,19). Furthermore, CSN5 and CSN6 facilitate degradation of p53 via stabilization of MDM2 (11,20). CSN5 also mediates nuclear export of p53 via phosphorylation of p53 at Thr155 (21,22). Interestingly, CSN6 was reported to stabilize several important oncogenic proteins such as β-catenin (13), Myc (23) and EGFR (12), thus promote cancer development and progression.
Intriguingly, recent studies also demonstrate that CSN5 is also implicated in mammalian innate immunity (24) and mediates the crosstalk between cancer cells and surrounding inflammatory niche. In macrophage, CSN5 is required for both Toll-like receptor (TLR) and reactive oxygen species-mediated deneddylation of Cul3. CSN5 KO potentiated NF-κB signaling and proinflammatory cytokine expression in macrophages from mouse model of atherosclerosis (25). CSN5 is involved in activation of NF-kB and TLR-mediated MAPK signal pathways in macrophage, and release of inflammatory cytokines (26).
Although CSN5 and CSN6 are frequently overexpressed in a variety of cancer types, it is unclear whether free CSN subunits or holo-complex are up-regulated in human cancers. As CSN5 can exist as free form or CSN-associated form, and CSN6 can exist either in the full or small CSN complex, we thus chose to examine the expression patterns of these subunits in LUAD specimens and determined their clinical significance. Our results demonstrated that CSN5, but not CSN6, was up-regulated in LUAD, and CSN5 is a potential prognostic predictor and therapeutic target for this lethal disease.
Methods
Patient information and tumor specimens
This study has been approved by the Institutional Review Board of the First Hospital of Foshan City, Guangdong, China (Approval Number L2014-01). The formalin fixed and paraffin embedded (FFPE) tumor specimens were retrospectively obtained from 59 patients who were diagnosed as lung adenocarcinoma (LUAD) and had undergone surgical resection in Department of Thoracic Surgery, The First Hospital of Foshan City, Guangdong, China. None of the patients received any anticancer therapies before surgery. Following resection, the tumor specimens were evaluated by pathologists to determine tumor-node-metastasis (TNM) stage according to the Union Internationale Contre le Cancer (UICC-7) staging system for lung cancer.
Immunohistochemical (IHC) staining
IHC staining was carried out using the standard streptavidin-perosidase method with DAB detection kit according to the manufacturer’s instructions. In brief, the FFPE tissue specimens were cut into 4 µm thick tissue sections onto glass slides then de-waxed in xylene and rehydrated through graded alcohols and water. Heat-induced Antigen retrieval was carried out in boiling Tris-EDTA buffer (pH 9.0) using a microwave oven. Endogenous peroxidase activity was blocked by incubating the sections in 3% hydrogen peroxide. After blocking with 10% normal goat serum in PBS, the tissue sections were incubated with primary antibody against human CSN5 or CSN6 (1:100, ABclonal Technology, Wuhan, China) overnight at 4 °C. The Detection System Peroxidase/DAB, Rabbit/Mouse kit (OriGene Technologies, Beijing, China) was applied to determine immunoreactivity according to the manufacturer’s instructions. Sections were then counterstained with hematoxylin and mounted in an aqueous mounting medium. The slides were scanned using a whole slide scanner (UNIC Healthcare) and image was captured using iViewer 6.2.
Immunostaining analysis
Analysis of immunostaining was performed by two independent pathologists by measuring the percentage of positive cells and staining intensity. The interobservers’ reproducibility of scoring immunostaining was assessed by Kappa statistic. The percentage of immunoreactive cells was scored as 1, positive cells 0–20%; 2, positive cells 21–50%; 3, positive cells 51–75%; and 4 positive cells 76–100%. The staining intensities were graded as 1, negative; 2, weak; 3, moderate; and 4, strong. The staining Index is scored by multiplying the percentage of positive cells by the intensity (I). A consensus was reached for intensity by taking the mean if scores were within 20% of each other; otherwise, images were reviewed and a consensus reached. Cases with a score >6 were considered as high expression in this study.
Cell culture
Human lung cancer H1299 and A549 cells were obtained from ATCC and routinely grown in DMEM medium supplemented with 10% FBS and antibiotics.
Construction of lentiviral-based vector and short hairpin RNA (shRNA), lentivirus production and infection
To construct the lentivirus-based CSN5 overexpression plasmid, cDNA was synthesized using total RNA extracted from normal lung tissue. The coding sequence for human CSN5 with FLAG-tag was obtained through amplification using normal lung tissue cDNA and inserted into the NheI and NotI sites of pCDH-CMV-MCS -EF1-puro (System Biosciences, USA). To generate lentiviral shRNA targeting CSN5, the oligonucleotides were annealed and inserted into HpaI and XhoI site of pLenti-mU6-CMV-GFP (Forevergen, Guangzhou, China). The sequences of oligonucleotides were as follows: shCSN5-1 sense, 5'-TGTCTCAGGTTATTAAGGATAA TTCAA GAGATTATCCTTAATAACCTGAGACTTTTTTC-3'; shCSN5-1 antisense, 5'-tcgagAAAAAAGTCT CAGGTTATTAAGGATAATCTCTTGAATTATCCTTAATAACCTGA- GACA-3'; shCSN5-2 sense, 5'-TCAGTCTCTGAGAAGTACTTTATTCAAGAGATAAAG TACTTCTCAG AGACTGTTTTTTC-3'; shCSN5-2 antisense, 5'-tcgagAAAAAACAGTCTCTG AGAAGTACTTTATCTCT TGAATAAAGTACTTCTCAGAGA -CTGA-3'. The sequences of the constructs were verified by sanger sequencing. To generate lentiviral particles, CSN5 expression or shRNA plasmid was cotransfected with psPAX2 and pMD2.G into 293T cells using lipofectamine 2000 (Life technologies, USA). The lentiviral particles were then used to infect lung cancer H1299 and A549 cells in the growth medium containing 8 µg/mL polybrene. The expression of CSN5 was confirmed by western blotting with the anti-FLAG tag antibody or anti-CSN5 antibody.
MTT Cell proliferation assay
Lung cancer cells infected with lentiviral CSN5 shRNA or stably overexpressing FLAG-tagged CSN5 were grown in 96-well plates. After 3 days, the cells were incubated with MTT (0.5 mg/mL) at 37 °C for 3 hours. One hundred µL DMSO was added into each well to dissolve formazan crystals and the absorbance was recorded at 570 nm.
Western blotting
Total cellular proteins were extracted by RIPA lysis buffer containing protease inhibitors. Protein concentrations were measured using Bradford assay (Bio-Rad), with BSA as the standard. Equal amounts of protein extracts from all samples were applied to SDS-PAGE and then transferred to PVDF membrane (Millipore). The membrane was blocked in Tris Buffered Saline buffer (TBS) containing 5% nonfat milk and 0.1% Tween-20 for 1 hour at room temperature followed by incubation with the primary antibody at 4 °C overnight. After being washed, membranes were incubated with the corresponding second antibodies. The signals were visualized by the enhanced chemiluminescence system (Bio-Rad) according to the manufacturer’s instructions.
Statistical analysis
Statistical analyses were carried out using IBM SPSS statistics 19 and GraphPad Prism 6. The Pearson’s Х2 test was used to determine the correlations between protein expression and clinical variables. Kaplan-Meier survival curves with log-rank test were used to estimate and compare the overall survival (OS) of the patients with different expression levels of CSN5. The Cox regression model with ENTER method was used to analyze he prognostic significance of the expression of CSN5. P<0.05 was considered to be significant.
Results
The expression of CSN5 and CSN6 in tumor specimens of LUAD patients
To explore the role of CSN5 and CSN6 in development of LUAD, we first examined their expressions in LUAD tumor specimens using IHC staining. Kappa statistic indicated the good agreement between two pathologists for both CSN5 and CSN6 (Kappa =0.891 for CSN5 and 0.839 for CSN6). As shown in Figure 1A and B, IHC staining revealed that CSN5 and CSN6 exhibited distinct staining patterns. It has been reported that CSN-associated CSN5 is predominantly localized in nuclear, whereas free CSN5 appears to be both cytoplasmic and nuclear (2). Consistent to previous observations, CSN5 showed some cytoplasmic staining, but predominant nuclear staining. In contrast, CSN6 was reported to be either in the full or small CSN complex. The full CSN complex was reported to localize in the nucleus, whereas the small CSN complex was localized in the cytoplasm (18,27). We found that CSN6 displayed prominent cytoplasmic distribution in LUAD (Figure 1B). Of note, as shown in Figure 1C, the expression levels of CSN5 were significantly higher in the tumor islet than those in adjacent normal tissues and in surrounding stromal cells. CSN5 was highly expressed in 66.1% (39/59) patients with a cut point as IHC Score >6. Conversely, CSN6 was barely detected (7/59, 11.9%) in the tumor tissues, and there was no significant difference in the CSN6 expression levels between tumor tissues and adjacent normal tissues (Figure 1C, right panel) even though the expression levels of CSN6 and CSN5 were weakly correlated (Figure 1D, r=0.404).
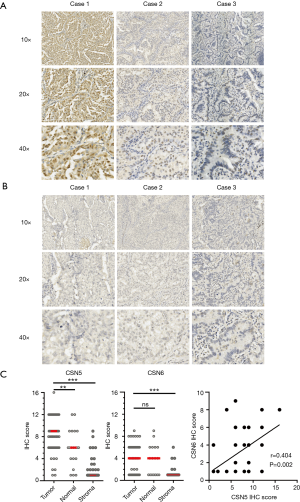
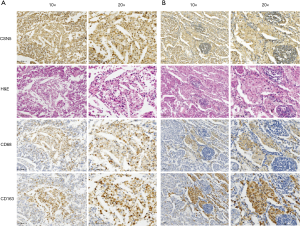
Interestingly, CSN5 staining was also positive in a subset of macrophages (CD68+ and CD163+) infiltrated in the tumor (Figure 2A) or adjacent to the tumor (Figure 2B).
The associations of CSN5 expression and clinical characteristics
Considering the low expression levels of CSN6, we thus focused on CSN5 in further analysis and experiments. We analyzed the relationship between CSN5 expression and clinicopathological parameters. As shown in Table 1, Pearson’s correlation analyses revealed that high CSN5 expression was associated with female, high TNM stage and N status. Moreover, Kaplan-Meier survival curve showed that higher levels of CSN5 were associated with worse progression-free survival (PFS) and OS of LUAD patients (Figure 3A).

Full table
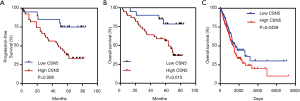
To validate the clinical significance of CSN5 in LUAD, we extracted the RNAseq data from the Cancer Genome Atlas (TCGA) and correlated the CSN5 mRNA levels with clinical outcomes of LUAD patients. Kaplan-Meier survival analysis confirmed that the higher levels of CSN5 mRNA were also associated with worse OS of LUAD patients (Figure 3B).
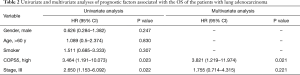
To further determine the prognostic value of CSN5, we performed Cox proportional hazard regression analysis. Univariate Cox regression analysis showed that TNM stage and CSN5 expression level were correlated with OS of the patients (Table 2). Further multivariate Cox regression analysis revealed that CSN5 alone was an independent risk factor for the clinical outcome of the LUAD patients (Table 2).
Full table
The role of CSN5 in the growth of lung cancer cells
In efforts to investigate the role of CSN5 in lung cancer, we modulated the expression of CSN5 in lung cancer H1299 and A549 cells via lentiviral-based overexpression vector or shRNA vector. As shown in Figure 4A and B, when the expression of CSN5 protein was effectively knocked down in H1299 and A549 cells, the cell proliferation assay showed that the growth of H1299 and A549 cells was strongly suppressed (Figure 4A and B, lower panel), whereas overexpression of CSN5 slightly promoted the growth of the lung cancer cells (Figure 4C,D). These results suggested that CSN5 silencing markedly suppressed lung cancer cells, and CSN5 was required for the growth of lung cancer cells.

Discussion
In the present study, we determined the expression patterns of CSN subunit CSN5 and its dimerization partner, CSN6, in the LUAD tumor specimens. The results showed that CSN5 was the major COP9 subunit that was preferentially expressed in the tumor cells, whereas its dimerization partner, CSN6 was barely detected in the tumor tissues of LUAD. Importantly, the expression levels of CSN5 were associated with TNM stage and clinical outcomes of LUAD patients, and further cellular study revealed that CSN5 inhibition could markedly suppress the growth of lung cancer cells. These results suggested that CSN5 serve as a potential prognostic predictor and therapeutic target for LUAD patients.
CSN5 shares sequence structural similarity with CSN6, and also forms heterodimer with CSN6. However, we found that CSN5 and CSN6 were not coordinately expressed in the LUAD tumor tissues even though their expression levels were weakly correlated: CSN5 was highly expressed in the tumor tissues, whereas CSN6 was hardly detective in the tumors. In fact, although the expressions of CSN subunits were investigated in various types of cancer, CSN subunits were seldom examined simultaneously in human cancers. It is unknown whether all CSN subunits or individual subunit were induced in human cancers. As CSN6 is integrated into the CSN, and it is unlikely to release from the holo-complex, whereas CSN5 is the only subunit which can leave and reenter the CSN complex. It has been reported that a large fraction of overexpressed CSN5 is found in free form which is localized in both cytoplasm and nuclear. Consistent to the previous studies, we found CSN5 showed some cytoplasmic staining, but predominant nuclear staining and was significantly upregulated in LUAD specimens. Thus, we speculate that free CSN5 may be the major form that is up-regulated in LUAD.
Importantly, recent evidence demonstrated that CSN5 possesses deubiquitination activity independent of CSN complex. It was reported that purified CSN5, which presumably lacks of deneddylation activity, modulates the deubiquitination and stabilization of PD-L1. These studies suggest that free CSN5 might contribute to carcinogenesis and tumor progression. Considering the importance of CSN in fundamental cellular processes, this finding also raises the possibility that we can develop specific inhibitor for monomeric or free CSN5 which could suppress cancer, without affecting the activity of CSN holo-complex. Indeed, several specific inhibitors of CSN5, such as CSN5i-3 (28), Thiolutin (29), Curcumin (30) and Azaindoles (31), have been identified and showed potential for anti-tumour therapy.
Emerging evidence suggests that CSN5 is implicated in chronic inflammation and immune response. As a downstream effector of NF-κB signaling, CSN5 in tumor cells is induced in response to inflammatory cytokine. As the immune cells in the tumor specimens are also surrounded by inflammatory milieu, thus it is not surprising that CSN5 was expressed in a proportional of macrophage infiltrated in or adjacent to the tumor islet. Interestingly, CSN5 is up-regulated in response to the inflammatory factor secreted by surrounding macrophage and stabilizes the immunosuppressive factor PD-L1 (8). CSN5 is also increased in human atherosclerotic arteries (25,32). However, the clinical and biological significance of CSN5 expression in immune cells and detailed mechanisms remain to be investigated.
Of note, there are some limitations in this study. First, the sample size of the cohort is relatively small, the finding in this study need to be validated in larger cohort even though we have validated the clinical significance of CSN5 in TCGA cohort. Second, although the expression patterns of CSN5 and CSN6 support the notion that free CSN5 is upregulated in LUAD and Inhibition of CSN5 is capable to suppress the growth of lung cancer cells, however, we did not examine the other subunits and we could not exclude the possibility that CSN5 was coordinately induced with the other subunits in LUAD.
In conclusion, this study demonstrates that CSN5 is the major COP9 subunit that is highly expressed in LUAD. CSN5 is also a critical regulator for the growth of lung cancer and represents an independent prognostic factor and a promising therapeutic target for LUAD patients.
Acknowledgements
Funding: This study was supported by the Natural Science Foundation of Guangdong Province (2016A030313721 and 2017A030313484).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study has been approved by the Institutional Review Board of the First Hospital of Foshan City, Guangdong, China (Approval Number L2014-01).
References
- Lingaraju GM, Bunker RD, Cavadini S, et al. Crystal structure of the human COP9 signalosome. Nature 2014;512:161-5. [Crossref] [PubMed]
- Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol 2003;19:261-86. [Crossref] [PubMed]
- Echalier A, Pan Y, Birol M, et al. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc Natl Acad Sci U S A 2013;110:1273-8. [Crossref] [PubMed]
- Birol M, Enchev RI, Padilla A, et al. Structural and biochemical characterization of the Cop9 signalosome CSN5/CSN6 heterodimer. PLoS One 2014;9:e105688. [Crossref] [PubMed]
- Kotiguda GG, Weinberg D, Dessau M, et al. The organization of a CSN5-containing subcomplex of the COP9 signalosome. J Biol Chem 2012;287:42031-41. [Crossref] [PubMed]
- Pick E, Golan A, Zimbler JZ, et al. The minimal deneddylase core of the COP9 signalosome excludes the Csn6 MPN- domain. PLoS One 2012;7:e43980. [Crossref] [PubMed]
- Liu Y, Shah SV, Xiang X, et al. COP9-associated CSN5 regulates exosomal protein deubiquitination and sorting. Am J Pathol 2009;174:1415-25. [Crossref] [PubMed]
- Lim SO, Li CW, Xia W, et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016;30:925-39. [Crossref] [PubMed]
- Lee MH, Zhao R, Phan L, et al. Roles of COP9 signalosome in cancer. Cell Cycle 2011;10:3057-66. [Crossref] [PubMed]
- Pan Y, Yang H, Claret FX. Emerging roles of Jab1/CSN5 in DNA damage response, DNA repair, and cancer. Cancer Biol Ther 2014;15:256-62. [Crossref] [PubMed]
- Zhao R, Yeung SC, Chen J, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest 2011;121:851-65. [Crossref] [PubMed]
- Hou J, Deng Q, Zhou J, et al. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene 2017;36:1134-44. [Crossref] [PubMed]
- Fang L, Lu W, Choi HH, et al. ERK2-Dependent Phosphorylation of CSN6 Is Critical in Colorectal Cancer Development. Cancer Cell 2015;28:183-97. [Crossref] [PubMed]
- Chen B, Zhao R, Su CH, et al. CDK inhibitor p57 (Kip2) is negatively regulated by COP9 signalosome subunit 6. Cell Cycle 2012;11:4633-41. [Crossref] [PubMed]
- Guo H, Jing L, Cheng Y, et al. Down-regulation of the cyclin-dependent kinase inhibitor p57 is mediated by Jab1/Csn5 in hepatocarcinogenesis. Hepatology 2016;63:898-913. [Crossref] [PubMed]
- Claret FX, Hibi M, Dhut S, et al. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 1996;383:453-7. [Crossref] [PubMed]
- Shin J, Phan L, Chen J, et al. CSN6 positively regulates c-Jun in a MEKK1-dependent manner. Cell Cycle 2015;14:3079-87. [Crossref] [PubMed]
- Tomoda K, Kubota Y, Arata Y, et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem 2002;277:2302-10. [Crossref] [PubMed]
- Choi HH, Guma S, Fang L, et al. Regulating the stability and localization of CDK inhibitor p27(Kip1) via CSN6-COP1 axis. Cell Cycle 2015;14:2265-73. [Crossref] [PubMed]
- Zhang XC, Chen J, Su CH, et al. Roles for CSN5 in control of p53/MDM2 activities. J Cell Biochem 2008;103:1219-30. [Crossref] [PubMed]
- Lee EW, Oh W, Song HP, et al. Phosphorylation of p53 at threonine 155 is required for Jab1-mediated nuclear export of p53. BMB Rep 2017;50:373-8. [Crossref] [PubMed]
- Bech-Otschir D, Kraft R, Huang X, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J 2001;20:1630-9. [Crossref] [PubMed]
- Chen J, Shin JH, Zhao R, et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun 2014;5:5384. [Crossref] [PubMed]
- Deng Z, Pardi R, Cheadle W, et al. Plant homologue constitutive photomorphogenesis 9 (COP9) signalosome subunit CSN5 regulates innate immune responses in macrophages. Blood 2011;117:4796-804. [Crossref] [PubMed]
- Asare Y, Ommer M, Azombo FA, et al. Inhibition of atherogenesis by the COP9 signalosome subunit 5 in vivo. Proc Natl Acad Sci U S A 2017;114:E2766-E75. [Crossref] [PubMed]
- Schwarz A, Bonaterra GA, Schwarzbach H, et al. Oxidized LDL-induced JAB1 influences NF-kappaB independent inflammatory signaling in human macrophages during foam cell formation. J Biomed Sci 2017;24:12. [Crossref] [PubMed]
- Adler AS, Littlepage LE, Lin M, et al. CSN5 isopeptidase activity links COP9 signalosome activation to breast cancer progression. Cancer Res 2008;68:506-15. [Crossref] [PubMed]
- Schlierf A, Altmann E, Quancard J, et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat Commun 2016;7:13166. [Crossref] [PubMed]
- Lauinger L, Li J, Shostak A, et al. Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat Chem Biol 2017;13:709-14. [Crossref] [PubMed]
- Uhle S, Medalia O, Waldron R, et al. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J 2003;22:1302-12. [Crossref] [PubMed]
- Altmann E, Erbel P, Renatus M, et al. Azaindoles as Zinc-Binding Small-Molecule Inhibitors of the JAMM Protease CSN5. Angew Chem Int Ed Engl 2017;56:1294-7. [Crossref] [PubMed]
- Asare Y, Shagdarsuren E, Schmid JA, et al. Endothelial CSN5 impairs NF-kappaB activation and monocyte adhesion to endothelial cells and is highly expressed in human atherosclerotic lesions. Thromb Haemost 2013;110:141-52. [Crossref] [PubMed]


