Deep learning and medical imaging
We live in an exciting period, which many call the Information Age, signifying the fact that the amount of information in the world is growing exponentially. This surge of information is of course also true for visual data, with a massive growth in photo and video images. More than a trillion photos are taken annually and billions are shared online, and every minute up to 300 hours of video are uploaded to YouTube alone. The healthcare system is also part of the exponential increase in textual and visual digital information (1) and the doubling time of medical knowledge is estimated to increase from 7 years in 1980 to 73 days in 2020 (1,2).
While uncategorized data goes to waste, it is already not humanely possible to categorize the exponentially increasing data, and efforts are continually made to find machine based solutions for data categorization. A major breakthrough in image classification (3) was the contribution of Krizhevsky et al. (4) to the ImageNet challenge in December 2012. The ImageNet database (5) is a large visual database designed for use in visual object recognition software research. In the 2012 ImageNet challenge, 1.3 million high-resolution images set into 1,000 different classes (such as cats, birds, cars etc.) were used to evaluate competing algorithms in the task of image classification (i.e., labeling an image of a cat as “cat”). Krizhevsky et al. proposed a Convolutional Neural Network (CNN) deep learning algorithm, termed AlexNet, which showed a remarkable 15% top-five classification error (4). Since then, each year, newer, deeper, CNN algorithms have shown improved error rates, achieving near human performance (6,7). The winners of the 2017 challenge, Hu et al., with their SENet algorithm, achieved a truly remarkable error rate of 2.25% (8).
Automatic image analysis (including medical images) is termed computer vision, which is an interdisciplinary field that deals with how computers can gain understanding of digital images or videos. Computer vision seeks to automate tasks that biological visual systems can do. Common image analysis tasks include classification, detection and segmentation (3). In the classification task the algorithm aims to classify images into two or more classes. Examples include classification of lung nodules into benign or malignant or classification of images into those containing cats and dogs. In the detection task the algorithm aims to localize structures in 2D or 3D space—for example, detection of lung nodules or liver metastases on CT images. In the segmentation task the algorithm tries to provide a pixel wise delineation of an organ or pathology—for example, segmentation of the surface of lung, kidney, spleen or tumors on CT, ultrasound or MRI images.
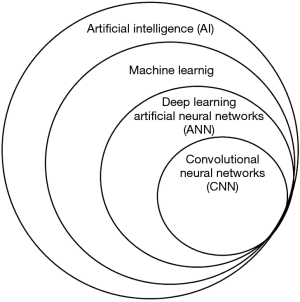
CNN algorithms, which are algorithms used to solve computer vision tasks, are part of a hierarchy of terms under the “umbrella” of Artificial Intelligence (AI) (9). Figure 1 presents a Venn diagram schematic representation of this hierarchy of terms. AI is a broad term that describes the computer science field that is devoted for creating algorithms that solve problems which usually require human intelligence. Machine learning is a subclass of AI that gives computers the ability to learn without being explicitly programmed. In classic machine learning, human experts try to choose imaging features that appear to best represent the visual data (for example, Hounsfield units histograms or the acuteness of masses) and statistical techniques are used to classify the data based on such features. Deep learning, a subtype of machine learning, is a type of representation learning in which no feature selection is used. Instead, the algorithm learns on its own which features are best for classifying the data. With enough training data, representation learning could potentially outperform hand-engineered features (9).
Artificial Neural Networks (ANN) forms the basis of most deep learning methods. ANNs are loosely based on the hypothesis of how biological neural networks operate: data enters the neuron’s dendrites, some kind of “mathematical function” is performed in the neuron’s body, and the result is outputted through the axon. The dendrites and axon terminals of many neurons interconnect in the brain’s cortex to create the biological neural network.
A schematic representation of ANN is presented in Figure 2. The first layer is the input layer (for example, intensity values of each pixel can represent each input). The black arrows in Figure 2 are sometimes termed weights, dendrites or parameters. Each weight is assigned a random number at initialization. Each input is multiplied by its corresponding weight, and the results are summed for each “neuron” in the network (∑). In this model “neurons” are just simple mathematical functions, mostly the ReLU function (x=0 for ∑≤0, x=∑ for ∑>0). The result of each neuron (x) is passed on and multiplied again by its corresponding weight. This process is repeated in all the neurons in the network, and a final result is outputted in the output neuron.
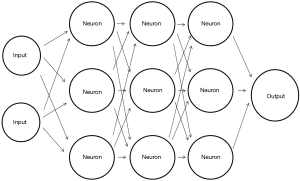
The true power of ANNs comes from the large amount of neurons which are arranged in interconnected deep hidden layers which gives the network its computational strength. Another key concept of ANN is backpropagation which is the process of network optimization. A loss function estimates the error, which is the difference between the output label and the desired label (e.g., did the network classify an image of a cat as a cat). Based on the estimation of error, the algorithm backpropagates through the network and small changes are made to the weights in order to reduce loss. Repeated iterations of forwarding all data inputs and backpropagating through the network, ultimately leads to an optimized network.
CNN are a subtype of ANN. Pioneered by LeCun et al. in 1990 (10), CNN were originally used to classify digits, and were applied to recognize hand-written numbers on bank checks, but as stated, the big breakthrough of CNN was made in 2012 in the ImageNet challenge (4).
CNN were inspired by the work of Hubel and Wiesel who won the Nobel prize for their understanding of the visual cortex pathways of cats (11). Specifically, Hubel and Wiesel observed that the visual process is a hierarchical rather than holistic process: neurons (termed “simple cells”) at the beginning of the visual process respond to simple features—lines at different angles. Higher level neurons (termed “complex cells” and “hyper-complex cells”) respond to complex combinations of simple features, for example, two lines at right angles to each other. CNN try to replicate this hierarchy of features. CNN operates by the process of convolution—point wise multiplication (dot product) of small matrices of weights across the entire image to create corresponding neuronal maps. The usual size of a filter is 3×3, 5×5 or 7×7 pixels. The process of matrix convolution is repeated on the constructed neural maps, to create new hidden deep layers of neural maps. Network optimization using loss estimation and backpropagation are conducted on the constructed neural network similarly to ANN.
The current paper, “Predicting malignancy of pulmonary ground-glass nodules (GGN) and their invasiveness by random forest”, by Mei and colleagues, presents a machine learning approach (see Figure 1) for the task of classifying pulmonary GGN. Mei et al. evaluated several classic machine learning algorithms (random forests, logistic regression, decision tree, support vector machine and AdaBoosting) for classifying CT images of pulmonary GGN from 1,177 patients. As in other classic machine learning algorithms, hand crafted imaging features were selected to represent the data. Two binary classification tasks were evaluated: (I) benign versus malignant status of GGNs; (II) non-invasive versus invasive status for the malignant nodules. The best machine learning algorithm was found to be random forests, with 95.1% accuracy to predict the malignancy of GGNs and 83.0% accuracy to predict the invasiveness of the malignant GGNs. As the authors state in the discussion, it will now be of interest to use this GGNs data-set to compare the classic machine learning algorithms with deep learning algorithms (mainly, CNN), which unlike machine learning algorithms, do not require the explicit use of radiologic features, but instead utilize the imaging information directly from the CT images.
Deep learning image analysis of chest radiology has been an active subject of research for the last two years. Research topics include, among others, classification of lung nodules into benign and malignant ones on CT scans (12-18), detection of lung nodules on CT scans (19,20), identification of interstitial lung disease on CT scans (21) and classification of mediastinal lymph nodes into benign and malignant on PET/CT (22).
While AI medical imaging research, such as in the studies described above, is rapidly increasing, and commercial applications are beginning to emerge, it is important to understand that although radiology is a somewhat large financial market, the driving force for AI development is much larger. Huge financial markets such as internet giants like Google, Facebook, Baidu and Alibaba, self-driving cars, robotics and many others, are the driving force for the financial and ingenuity investment in the development of both the software and the hardware aspects of AI. New developments in machine learning, and more specifically deep learning, will likely have major implications on human society in the near future, and this advancement has already been termed by some, the fourth industrial revolution (23). In a summary of the Intersociety Summer Conference of the American College of Radiology (ACR) it has been stated that “Data science will change radiology practice as we know it more than anything since Roentgen, and it will happen more quickly than we expect.. pretending this will not disrupt radiology is naive” (1).
Some radiology experts emphasize the importance of remaining optimistic about future opportunities of data science (1), as well as about the active role of radiologists in the AI revolution, a role that entails engagement rather than replacement (24,25). The future is unknown, the AI revolution has just recently started, and as humans we struggle in estimating exponential processes. It is important for medical institutions to invest in the understanding of this new technology, in order to be best prepared for the upcoming changes.
Acknowledgements
None.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Kruskal JB, Berkowitz S, Geis JR, et al. Big Data and Machine Learning-Strategies for Driving This Bus: A Summary of the 2016 Intersociety Summer Conference. J Am Coll Radiol 2017;14:811-7. [Crossref] [PubMed]
- Densen P. Challenges and opportunities facing medical education. Trans Am Clin Climatol Assoc 2011;122:48-58. [PubMed]
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Proceedings of the 25th International Conference on Neural Information Processing Systems - Volume 1. Lake Tahoe, Nevada, Curran Associates Inc.; 2012:1097-105.
- Fei-Fei L, Deng J, Li K. ImageNet: Constructing a large-scale image database. Journal of Vision 2009;9:1037. [Crossref]
- Szegedy C, Liu W, Jia Y, et al. Going Deeper with Convolutions. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 2015:1-9.
- He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 2016.
- Hu J, Shen L, Sun G. Squeeze-and-Excitation Networks. arXiv preprint arXiv:1709.01507, 2017.
- Chartrand G, Cheng PM, Vorontsov E, et al. Deep Learning: A Primer for Radiologists. Radiographics 2017;37:2113-31. [Crossref] [PubMed]
- LeCun, Boser B, Denker JS, et al. Handwritten digit recognition with a back-propagation network. In: David ST. editor. Advances in neural information processing systems 2. Burlington: Morgan Kaufmann Publishers Inc.; 1990:396-404.
- Hubel DH, Wiesel TN. Receptive fields of single neurones in the cat's striate cortex. J Physiol 1959;148:574-91. [Crossref] [PubMed]
- Ciompi F, de Hoop B, van Riel SJ, et al. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out-of-the-box. Med Image Anal 2015;26:195-202. [Crossref] [PubMed]
- Nibali A, He Z, Wollersheim D. Pulmonary nodule classification with deep residual networks. Int J Comput Assist Radiol Surg 2017;12:1799-808. [Crossref] [PubMed]
- Song Q, Zhao L, Luo X, et al. Using Deep Learning for Classification of Lung Nodules on Computed Tomography Images. J Healthc Eng 2017;2017:8314740. [PubMed]
- Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. [Crossref] [PubMed]
- Hua KL, Hsu CH, Hidayati SC, et al. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015;8:2015-22. [PubMed]
- Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med 2017;89:530-9. [Crossref] [PubMed]
- Wang H, Zhao T, Li LC, et al. A hybrid CNN feature model for pulmonary nodule malignancy risk differentiation. J Xray Sci Technol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Wang C, Elazab A, Wu J, et al. Lung nodule classification using deep feature fusion in chest radiography. Comput Med Imaging Graph 2017;57:10-8. [Crossref] [PubMed]
- Setio AA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal 2017;42:1-13. [Crossref] [PubMed]
- Anthimopoulos M, Christodoulidis S, Ebner L, et al. Lung Pattern Classification for Interstitial Lung Diseases Using a Deep Convolutional Neural Network. IEEE Trans Med Imaging 2016;35:1207-16. [Crossref] [PubMed]
- Wang H, Zhou Z, Li Y, et al. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from (18)F-FDG PET/CT images. EJNMMI Res 2017;7:11. [Crossref] [PubMed]
- Hricak H. 2016 New Horizons Lecture: Beyond Imaging-Radiology of Tomorrow. Radiology 2018;286:764-75. [Crossref] [PubMed]
- Kohli M, Prevedello LM, Filice RW, et al. Implementing Machine Learning in Radiology Practice and Research. AJR Am J Roentgenol 2017;208:754-60. [Crossref] [PubMed]
- Dreyer KJ, Geis JR. When Machines Think: Radiology's Next Frontier. Radiology 2017;285:713-8. [Crossref] [PubMed]


