Automatic detection of mycobacterium tuberculosis using artificial intelligence
Introduction
Tuberculosis (TB), a chronic infectious disease caused by mycobacterium TB, is a global issue that seriously endangers public health. In recent years, TB is becoming resurgence in worldwide after had been controlled in a large extent for many years. China is reported to have the second highest incidence rate of TB worldwide. According to the fifth national epidemiological sampling survey of TB in 2010, the prevalence of active TB in people aged 15 and over was 459/100,000. Prevention and treatment of TB have always been important tasks for medical professionals (1).
Pathology is one of the most important means for diagnosing TB in clinical practice. To confirm TB as the diagnosis, finding specially stained TB bacilli under a microscope is critical, in addition to identifying a set of morphologic changes in the surrounding tissue. Because of the very small size (less than 1µm in diameter) of the bacilli, to look for and identify them under the microscope requires use of high-power fields, which provide a rather limited visual area over a whole tissue section. Besides, the number of bacilli is usually small. Therefore, it is a time-consuming and strenuous work even for experienced pathologists, and this strenuosity often leads to low detection rate and false diagnoses. In order to improve the efficiency and sensitivity of the detection of TB bacilli, several new techniques have been developed, including PCR and RNA scope, but so far none of them have proved to be reliable and accepted widely (2).
In recent years, with the rapid development of information technology and the growing attention to interdisciplinary practices, artificial intelligence (AI) has become a new area of interest for medical professionals. The term first appeared in the Dartmouth Summer Research Project on Artificial Intelligence in 1956 (3). After 50 years of development and study, especially after recent breakthroughs on convolutional neural networks (4), AI has shown some amazing results on performing human-level tasks. For instance, AlexNet, a neural network model, achieved a great success in the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) in 2012 (5). In many other studies and challenges, AI has also demonstrated the capability to handle medical tasks.
Compared to the complex and varied shape of cells and tissues, the morphology of TB bacillus is relatively simple, resembling a thin rod with a length of roughly 4 µm and a diameter of 0.5–1.0 µm. The waxy lipid in the cell wall of the bacilli appears purple red after acid-fast staining, showing high contrast to the blue background (6). Detecting bacilli with such morphology and color upon staining is specific to the diagnosis of TB. In this paper, we present a novel method to automatically detect stained TB bacilli using a convolutional neural network model. We show that our method is highly competitive in terms of sensitivity and specificity, while being naturally consistent and efficient.
Methods
A total of 246 cases were collected from the Department of Pathology, Peking University First Hospital during January 2016 to June 2017. All of them were tissue samples and treated by acid-fast stain, of which 45 cases were selected as training set and 201 cases were selected as testing set.
All specimens were fixed, embedded, sectioned, and stained by acid-fast stain strictly following the standard protocols. Slides were reviewed, and the results were determined by two pathologists separately. Consensus was reached on inconsistent cases after discussion.
All slides were scanned using a KF-PRO-005 Digital Section Scanner (Ningbo Jiangfeng Bio-information Technology Co., Ltd., Ningbo, China). Pathologists provided 30 positive samples, containing a large number of TB bacilli, and 15 negative samples (where no signs of TB bacilli were found). The bacilli in the positive samples were labeled using the ASAP software. First, we sampled 96,530 small patches at 40× magnification level from labeled areas in the whole slide images as positive samples. We then performed further data augmentation, including rotation, mirroring, and position shifting, and eventually obtained a set of 578,191 patches with bacilli. Similarly, 2,510,307 negative samples were collected outside the bacilli regions. Our training set thus contained over 3 million samples.
We adopt a convolutional neural network model pre-trained on the CIFAR-10 dataset as our classification model, named tuberculosis artificial intelligence (TB-AI). Input to the network were 32×32 pixel patches from positive and negative regions of the WSI and the classification model was trained to discriminate between the positive and negative patches. We used mini-batch stochastic gradient descent algorithm as the optimization method, with a batch size of 512. The initial learning rate was set to 0.05 and was multiplied by 0.1 after every 5 epochs. The model was trained for 25 epochs in total. The 32×32 pixel patches were generated from the test slides using a sliding window method with a stride of 16. They were then fed into the trained TB-AI to generate a probability map for each slide. Patches with bacillus probability >0.5 were considered positive. A slide would be predicted positive if it contained at least one positive patch.
The diagnostic results made by TB-AI were compared to those made by pathologists. We analyzed inconsistencies between the results, and modified the protocol to get more accurate predictions. Step above was repeated
Results
Before the test, 108 out of 201 cases were diagnosed as positive by pathologists, and the remaining 93 cases were negative.
Trained TB-AI were run on the test data twice. The results are shown in Table 1. In the first run, the diagnoses of 154 cases (76.6%) made by TB-AI were consistent with those made by the pathologists, including 93 positive cases (46.2%) and 61 negative cases (30.3%). The diagnoses of the remaining 47 cases (23.4%) made by TB-AI were different from those made by the pathologists, of which 15 cases (7.5%) were diagnosed positive by pathologists but negative by TB-AI and 32 cases (15.9%) were diagnosed negative by pathologists but positive by TB-AI. In the second run, 28 more samples were diagnosed accurately, including 20 positive cases and 8 negative cases. With careful modifications to the model, we improved the consistency rate by almost 14% (Table 1).
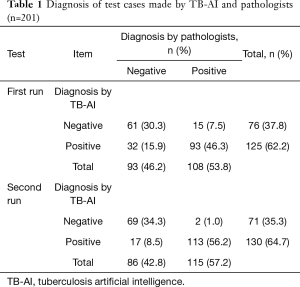
Full table
Discussion
Development of computer-aided diagnosis (CAD) systems to automatically detect acid-fast stained bacilli began in the 1990s. These computer programs often rely on identifying certain chromatic or structural patterns that are pre-defined by medical experts and engineers. Although bacillus carries a distinctive morphology and can be easily identified by trained pathologists, it’s difficult to parameterize the morphological features and precisely define the range within which they would vary. Therefore, these feature-based methods usually do not generalize well. CNN, a newly developed set of mathematical models, is able to simulate the work process of biological nervous systems and automatically learns the features that in traditional algorithms were hand-engineered, thus requiring relatively little pre-processing (7). This methodology is similar to how pathologists learn to recognize TB bacilli under the microscope, and hence should theoretically work well in our task. Our main challenge was that our targets were extremely small compared to the whole slide image: at 40× magnification, the size of a TB bacillus was only around 20×4 pixels. As a result, how to establish a neural network model that would apply to small objects became the key point in our study. Among the existing models, Google’s CIFAR-10 is designed to handle micro-images at a size of 32×32 pixels, which is very close to that of TB bacillus. In addition, it’s able to process the images at a very high speed (8). Based on CIFAR-10, a unique CNN model specifically targeting TB-AI was established.
The 47 inconsistent cases during the first run were reviewed by two pathologists both via microscope and digital slides. There are 7 positive cases missed by pathologists and 8 by TB-AI, in all of which existence of TB bacilli was confirmed after the review process (Figure 1). In 6 cases, dye residues were misdiagnosed as bacilli by TB-AI (Figure 2). In 20 cases, TB-AI identified bacilli located outside of tissues or on the surface of normal tissues, which were later ruled out by pathologists and regarded as contaminant bacilli (Figure 3). In the last 6 cases TB bacilli were found under the microscope, but neither pathologists nor TB-AI were able to identify them on the corresponding digital images due to the limited image quality of the scanned slides (Figure 4).

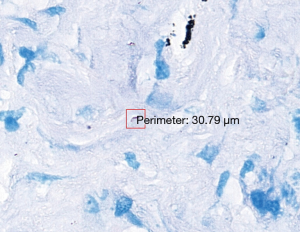
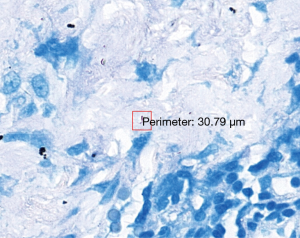

Based on the analysis above the protocol was improved as follows. First, the diagnosis mistakes in the test data were corrected by pathologists. Second, the scanner was reset to minimize compression losses and to increase the precision of the auto-focus program, hence improving the quality of the digital slides. Third, the training set was optimized by correcting mislabeled data and improving labeling accuracy. We also performed class balancing by expanding rarer samples. Fourth, to remedy the inadequacy of the diversity and coverage of negative samples, we extended the sampling area to cover more regions on the digital slides. Fifth, a random forest classifier was added to the framework so that outputs from the CNN would be filtered a second time to further improve specificity.
The general practice in machine learning prohibits using the test dataset a second time such that the model can remain unbiased. In our case, however, we found errors in the ground truth label during the first run, which had made the second run necessary. Yet we did not tune our model to make it biased towards the test data, and hence our model should remain neutral. In the second run, TB-AI missed 2 positive cases (1.0%) and misdiagnosed 17 negative cases (8.5%) where it miscounted contaminant bacilli towards pathogenic bacilli. With the diagnosis made by pathologists jointly via microscope and digital slides as the golden standard, the sensitivity and specificity of TB bacilli detection using TB-AI was 97.94% and 83.65% respectively.
Conclusions
We have developed an efficient and reliable framework for automatic TB bacilli detection based on deep learning and machine learning algorithms. We have shown that TB-AI demonstrates high sensitivity for the recognition of bacilli, but so far it is still relatively weak when it comes to distinguishing contaminant bacilli from pathogenic bacilli. Distinguishing pathogenic bacilli from contaminant bacilli depends on identifying the morphological changes caused by certain histological reactions, i.e., caseous necrosis, granuloma and inflammation. Currently, these knowledges have not been learned by TB-AI, and hence it is still necessary to have a pathologist to confirm the diagnosis results made by this system. TB-AI, a framework for automatically detecting acid-fast stained TB bacilli, shows a high sensitivity and moderate specificity. It is a promising method to relieve pathologists from such heavy duties as hunting for bacilli under a microscope and decrease chances of false diagnosis. In practice, positive results given by TB-AI need to be confirmed by pathologists, while negative results should be reviewed to make sure that the digital slides are qualified.
Acknowledgements
Funding: Supported by Beijing Municipal Science and Technology Commission (No. Z181100001918007).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study has been approved by the ethics committee of Peking University First Hospital.
References
- Technical Guidance Group of the Fifth National TB Epidemiological Survey, The Office of the Fifth National TB Epidemiolological Survey. The fifth national tuberculosis epidemiological survey in 2010. Chinese Journal of Antituberculosis 2012;34:485-508.
- Travis WD, Colby TV, Koss MN, et al. editors. Non-Neoplastic Disorders of the Lower Respiratory Tract. Washington D. C., USA: American Registry of Pathology; 2002:582.
- Solomonoff RJ. The The time scale of artificial intelligence: Reflections on social effects. Human Systems Management 1985;5:149-53.
- Goodfellow I, Bengio Y, Courville A, editors. Deep Learning. Boston, USA: MIT Press; 2016.
- Olga R, Deng Jia, Hao S, et al. ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision 2015;115:211-52. [Crossref]
- Suvarna KS, Christopher L, Bancroft JD. Bancroft’s Theory and Practice of Histological Techniques. London, UK: Churchill Livingstone; 2012:296-7.
- Matsugu M, Mori K, Mitari Y, et al. Subject independent facial expression recognition with robust face detection using a convolutional neural network. Neural Netw 2003;16:555-9. [Crossref] [PubMed]
- Song C, Xu DY, Qin YB. Research on detached multiple convolutional neural network. Computer Engineering 2017;43:145-9.


