Combination immuno-oncology therapy with immune checkpoint blockers targeting PD-L1, PD-1 or CTLA4 and epigenetic drugs targeting MYC and immune evasion for precision medicine
Therapeutics targeting epigenetic regulators (1-3), immune modulators (4-6) and receptor tyrosine kinases (7-9) have emerged as promising drugs for cancer patients who are refractory to conventional chemotherapy, radiotherapy or surgery (Figure 1). These cutting-edge therapeutics, together with next-generation sequencing (NGS) technologies that produce bulk genomics-related data as commodities of clinical medicine, have led to a gradual shift toward personalized or precision medicine. Whole genome sequencing comprehensively detecting alterations in common and rare cancer-related genes and cancer gene panels detecting alterations in approximately 300 common cancer-related genes are representative NGS-based sequencing (10-12). In current practice, personalized medicine involves using NGS-based diagnostic tests to select patients and time points for targeted therapy; in the future, precision medicine will involve utilizing multiple layers of omics data to optimize benefit-risk balance for targeted therapy (10). Lung cancer is one of the most common malignancies in the world (13). Non-small cell lung cancer (NSCLC) accounts for the majority of lung cancer cases, and NSCLCs can be sub-classified into lung adenocarcinoma, lung squamous cell carcinoma and other types of cancer. Epidermal growth factor receptor (EGFR) inhibitors (afatinib, erlotinib, gefitinib and osimertinib) and immuno-oncology drugs (atezolizumab, nivolumab and pembrolizumab) are representative targeted therapeutics approved for the treatment of NSCLC patients (14-16). Because recurrence and adverse effects are unavoidable even when cutting-edge targeted therapies are administered, the use of a combination of different categories of drugs is a rational strategy to enhance the benefits and reduce the drawbacks of targeted therapy.
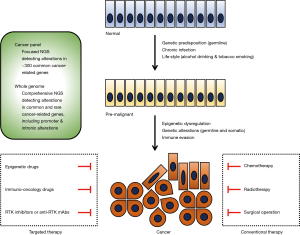
Epigenetic regulation of transcription and phenotypes involves chromatin-dependent mechanisms, such as methylation of genomic DNA and post-translational modification of histones (1-3). Because chromatin consists of genomic DNA wrapped around histone octamers, closed and open chromatin states are fine-tuned via DNA methylation and histone modification. In closed chromatin or heterochromatin, transcription factors cannot access genomic DNA; such chromatin is found in transcriptionally repressed regions, which are characterized by methylation of histone H3 lysine 9 (H3K9) and hypermethylation of genomic DNA. In contrast, in open chromatin or euchromatin, transcription factors can access genomic DNA; such chromatin is found in transcriptionally active regions, which are characterized by tri-methylation of H3K4 and H3K36 and hypomethylation of genomic DNA. CpG hypermethylation in the promoter regions of tumor suppressor genes induces the silencing of these genes, whereas genetic alterations in various epigenetic regulators, such as ASXL1, ASXL2, BAP1, DNMT3A, EZH2, IDH1, IDH2, MLL1, MLL3, NSD1, NSD2, NSD3, SMARCA4 and TET2, are involved in human carcinogenesis (1,2,17-23). Epigenetic dysregulation plays a pivotal role in human carcinogenesis. Bromodomain and extra-terminal (BET) inhibitors, DNA methyltransferase (DNMT) inhibitors, histone deacetylase (HDAC) inhibitors, histone lysine demethylase (KDM) inhibitors and histone lysine methyltransferase (KMT) inhibitors have been developed as drugs that target epigenetic regulators (1-3).
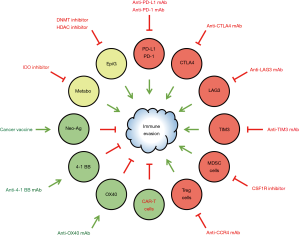
Immuno-oncology therapeutics are generally classified into inhibitors of immunosuppressive ligands or receptors (immune checkpoint blockers) and activators of immunostimulatory receptors (immunostimulatory agents) (4-6). CTLA4 (CD152), LAG3 (CD223), PD-1 (CD279/PDCD1), TIGIT and TIM3 (CD366/HAVCR2) are representative immunosuppressive receptors, whereas ICOS, TNFRSF4 (CD134/OX40), TNFRSF9 (CD137/4-1BB) and TNFRSF18 (CD357/GITR) are representative immunostimulatory receptors. Immunogenomic analyses have revealed that CTLA4, LAG3 and PD-1 are major immunosuppressive receptors in various types of human cancers and that TIGIT and TIM3 are additional immunosuppressive receptors that function in a tumor type-dependent manner (24). PD-1 ligand 1 (PD-L1/CD274) on cancer cells, macrophages, myeloid-derived suppressor cells and stromal cells in the tumor microenvironment interacts with PD-1 receptor on T cells to induce immune evasion via the inhibition of PI3K-AKT, RAS-MAPK and calcineurin-NFAT signaling. Blockade of this immunosuppressive signaling using monoclonal antibodies (mAbs) against PD-L1 or PD-1 induces the activation, differentiation and/or proliferation of tumor-infiltrating T cells via de-repression of the PI3K-AKT, RAS-MAPK and calcineurin-NFAT signaling cascades. Anti-PD-L1 mAbs (atezolizumab, avelumab and durvalumab), anti-PD-1 mAbs (nivolumab and pembrolizumab) and an anti-CTLA4 mAb (ipilimumab) are immune checkpoint blockers that have been approved for the treatment of cancer patients, whereas other immunomodulatory therapeutics that target 4-1BB, GITR, ICOS, LAG3, OX40, TIGIT and/or TIM3 are being accessed in clinical trials or preclinical research (4-6).
Durable partial or complete responses to immune checkpoint blockers, including pembrolizumab and nivolumab, have been observed in approximately 20% of NSCLC patients and 31-44% of melanoma patients (25); however, immune checkpoint blockade therapies have been associated with adverse events, including pneumonitis in 4.7% (43/915) of cancer patients (26) and anecdotal rapid progression of cancer or hyperprogressive disease in 9.2% (12/131) of cancer patients (27). Expression of PD-L1 ligand (28) and/or clonal neoantigen on tumor cells (29) and mismatch repair (MMR) deficiency (30) have been reported as biomarkers associated with preferable response to immune checkpoint blockers, whereas higher tumor burden (31), loss of neoantigen expression on tumor cells (32), escape mutations in the interferon signaling cascade in tumor cells (33) and upregulation of alternative immunosuppressive receptors on T cells (34) are biomarkers associated with poor response to immune checkpoint blockers. Comprehensive genotype-immunophenotype analyses should be performed in companion studies of clinical trials to generalize the mechanisms of durable response to, resistance against or severe adverse effects of immuno-oncology therapy in various types of human cancers. Further fine-tuning of the benefit-risk balances of immuno-oncology drugs is necessary for the application of these drugs as part of a precision oncology platform.
Recently, Topper et al. reported preclinical research on combination epigenetic therapy for NSCLC that evaluated (I) synergistic effects of a DNMT inhibitor (azacitidine) and an HDAC inhibitor (ITF-2357, MGCD-0103 or MS-275), especially the combination of azacitidine and ITF-2357, on cells from a panel of NSCLC cell lines and (II) epigenetic therapy-induced MYC downregulation, interferon signaling activation and CCL5, HLA-A and HLA-B upregulation in tumor cells, as well as downregulation of PD-1 and CTLA4 and upregulation of CCR7 in CD8+ tumor-infiltrating lymphocytes, in mouse models of lung cancer (35). Topper et al. postulated that depletion of the oncoprotein MYC and removal of immune evasion might explain the mechanisms by which epigenetic therapy involving DNMT and HDAC inhibitors produces anti-tumor effects. In contrast, Zheng et al. screened 97 approved oncology drugs and found that only the HDAC inhibitor romidepsin induces upregulation of the chemokines CCL5, CXCL9 and CXCL10 with T cell-attracting potential in mouse and human lung cancer cell lines (36). Zheng et al. demonstrated synergistic anti-tumor effects of romidepsin and anti-PD-1 mAb in mouse lung cancer models, and proposed upregulation of T cell-attracting chemokines and interferon-γ as mechanisms underlying the synergistic effects of the HDAC inhibitor and immune checkpoint blocker. These two reports clarified cross-talk between epigenetic dysregulation and immune evasion during lung cancer progression and emphasized a rational strategy involving the use of a combination of epigenetic drugs and immuno-oncology drugs for cancer therapy.
Immune checkpoint blockers can be combined with epigenetic therapeutics and other therapeutics (Figure 2), including alternative immune checkpoint blockers, cancer vaccines, conventional chemotherapy, immunostimulatory agents, macrophage inhibitors, metabolic modulators, natural killer cell inhibitors, radiotherapy, receptor tyrosine kinase inhibitors and regulatory T (Treg) cell inhibitors (6). For example, the following combination epigenetic immuno-oncology therapies are in clinical trials for cancer patients: an anti-PD-L1 mAb (atezolizumab) and a DNMT inhibitor (azacitidine) [ClinicalTrials.gov identifier: NCT02508870]; an anti-PD-L1 mAb (atezolizumab) and a DNMT inhibitor (guadecitabine) [ClinicalTrials.gov identifier: NCT03179943]; an anti-PD-1 mAb (pembrolizumab) and a DNMT inhibitor (CC-486) [ClinicalTrials.gov identifier: NCT02546986]; an anti-PD-1 mAb (pembrolizumab) and an HDAC inhibitor (entinostat) [ClinicalTrials.gov identifier: NCT02437136]; an anti-PD-1 mAb (pembrolizumab) and an HDAC inhibitor (romidepsin) [ClinicalTrials.gov identifier: NCT03278782]; and an anti-PD-1 mAb (pembrolizumab) and an HDAC inhibitor (vorinostat) [ClinicalTrials.gov identifier: NCT02638090].
Upregulation of MYC and downregulation of CCL5, CXCL9, CXCL10, HLA-A and/or HLA-B in pretreatment tumor samples might be predictive biomarkers of response to combination epigenetic immuno-oncology therapies, because DNMT and HDAC inhibitors synergistically revert immune evasion via MYC repression and reciprocal de-repression of CCL5, HLA-A and HLA-B (35) and HDAC inhibitors exert anti-tumor effects via de-repression of the chemokine CCL5, CXCL9 and CXCL10 (36). MYC is transcriptionally upregulated via genetic alterations in MYC, including gene rearrangement, gene amplification and focal amplification of the super-enhancer region (37), and activation of the WNT, Notch and other signaling pathways; it is post-translationally stabilized via PI3K-AKT and RAS-MAPK signaling activation (38). MYC activates its target genes to regulate a variety of cellular processes, such as proliferation, metabolism, survival and apoptosis, in a context-dependent manner (38,39). Taken together, these facts suggest that certain tumors with constitutive MYC overexpression might be resistant to epigenetic therapy-induced MYC repression and that epigenetic therapy-induced MYC repression might not always lead to upregulation of CCL5, HLA-A or HLA-B in tumor cells and the subsequent correction of immune evasion. Whole-genome sequencing and transcriptome analyses of tumor cells and immuno-phenotype analyses of the tumor microenvironment are necessary to precisely predict responders to epigenetic immuno-oncology therapies.
In conclusion, clinical trials and companion studies to develop the most effective combination immuno-oncology therapy and identify biomarkers that predict therapeutic benefits and risks of adverse effects are necessary to optimize the benefit-risk balance of precision medicine for cancer patients in the future.
Acknowledgements
This work was supported in part by a grant-in-aid from M. Katoh’s Fund for the Knowledge-Base Project.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Katoh M. Mutation spectra of histone methyltransferases with canonical SET domains and EZH2-targeted therapy. Epigenomics 2016;8:285-305. [Crossref] [PubMed]
- Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016;17:284-99. [Crossref] [PubMed]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487-500. [Crossref] [PubMed]
- Boussiotis VA. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med 2016;375:1767-78. [Crossref] [PubMed]
- Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. [Crossref] [PubMed]
- Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707-23. [Crossref] [PubMed]
- Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. [Crossref] [PubMed]
- Costa DB. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Transl Lung Cancer Res 2016;5:331-7. [Crossref] [PubMed]
- Katoh M. Therapeutics Targeting FGF Signaling Network in Human Diseases. Trends Pharmacol Sci 2016;37:1081-96. [Crossref] [PubMed]
- Katoh M. The integration of genomics testing and functional proteomics in the era of personalized medicine. Expert Rev Proteomics 2017;14:1055-8. [Crossref] [PubMed]
- Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023-31. [Crossref] [PubMed]
- Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. [Crossref] [PubMed]
- Torre LA, Siegel RL, Ward EM, et al. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol Biomarkers Prev 2016;25:16-27. [Crossref] [PubMed]
- Hirsch FR, Suda K, Wiens J, et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388:1012-24. [Crossref] [PubMed]
- Callahan MK, Postow MA, Wolchok JD, Targeting T. Cell Co-receptors for Cancer Therapy. Immunity 2016;44:1069-78. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Gelsi-Boyer V, Brecqueville M, Devillier R, et al. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol Oncol 2012;5:12. [Crossref] [PubMed]
- Katoh M. Functional and cancer genomics of ASXL family members. Br J Cancer 2013;109:299-306. [Crossref] [PubMed]
- Balasubramani A, Larjo A, Bassein JA, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun 2015;6:7307. [Crossref] [PubMed]
- Sahtoe DD, van Dijk WJ, Ekkebus R, et al. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun 2016;7:10292. [Crossref] [PubMed]
- Saeidi K. Myeloproliferative neoplasms: Current molecular biology and genetics. Crit Rev Oncol Hematol 2016;98:375-89. [Crossref] [PubMed]
- Chen HJ, Wei Z, Sun J, et al. A recellularized human colon model identifies cancer driver genes. Nat Biotechnol 2016;34:845-51. [Crossref] [PubMed]
- Guryanova OA, Lieu YK, Garrett-Bakelman FE, et al. Dnmt3a regulates myeloproliferation and liver-specific expansion of hematopoietic stem and progenitor cells. Leukemia 2016;30:1133-42. [Crossref] [PubMed]
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep 2017;18:248-62. [Crossref] [PubMed]
- Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275-87. [Crossref] [PubMed]
- Naidoo J, Wang X, Woo KM, et al. Pneumonitis in Patients Treated With Anti-Programmed Death-1/Programmed Death Ligand 1 Therapy. J Clin Oncol 2017;35:709-17. [Crossref] [PubMed]
- Champiat S, Dercle L, Ammari S, et al. Hyperprogressive Disease Is a New Pattern of Progression in Cancer Patients Treated by Anti-PD-1/PD-L1. Clin Cancer Res 2017;23:1920-28. [Crossref] [PubMed]
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Huang AC, Postow MA, Orlowski RJ, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60-5. [Crossref] [PubMed]
- Anagnostou V, Smith KN, Forde PM, et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov 2017;7:264-76. [Crossref] [PubMed]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov 2017;7:188-201. [Crossref] [PubMed]
- Koyama S, Akbay EA, Li YY, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun 2016;7:10501. [Crossref] [PubMed]
- Topper MJ, Vaz M, Chiappinelli KB, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell 2017;171:1284-300.e21. [Crossref] [PubMed]
- Zheng H, Zhao W, Yan C, et al. HDAC Inhibitors Enhance T-Cell Chemokine Expression and Augment Response to PD-1 Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res 2016;22:4119-32. [Crossref] [PubMed]
- Zhang X, Choi PS, Francis JM, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genet 2016;48:176-82. [Crossref] [PubMed]
- Kress TR, Sabò A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 2015;15:593-607. [Crossref] [PubMed]
- Stine ZE, Walton ZE, Altman BJ, et al. MYC, Metabolism, and Cancer. Cancer Discov 2015;5:1024-39. [Crossref] [PubMed]


