Current understanding of middle east respiratory syndrome coronavirus infection in human and animal models
Introduction
Since the discovery of first human coronavirus (HCoV), HCoVs have been studied for over 50 years (1). Overall six HCoVs have been identified including HCoV-229E and HCoV-OC43 (first identified in the 1960s), SARS-CoV (in 2003), HCoV-NL63 (in 2004), HCoV-HKU1 (in 2005) and Middle East respiratory syndrome coronavirus (MERS-CoV) (in 2012) (1-5). Zoonotic coronaviruses such as SARS-CoV (6) and MERS-CoV (7), crossed barrier and lead to epidemic in humans. MERS-CoV as the most recent novel coronavirus emerged in human can cause severe, life-threatening disease and is a potential threat to global public health and economy. Since 2012, MERS cases, including those transmitted from dromedary camels were reported to World Health Organization (WHO) almost every month. In May 2015, a returned traveler with MERS-CoV infection caused a significant outbreak in South Korea, which spread to 186 patients and over 16,000 people were quarantined (8). This outbreak raised the fear that a pandemic like SARS would reoccur. Here, we reviewed the epidemiology, animal model generations and most recent progress on vaccine and treatment developments against MERS-CoV.
Origin and evolution
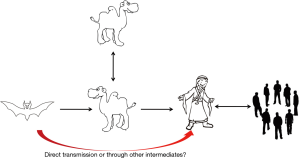
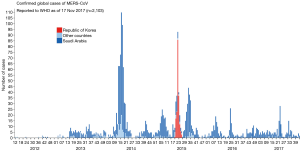
MERS-CoV was first emerged in Saudi Arabia in 2012. As of January2018, WHO has been notified of 2143 laboratory-confirmed cases including 750 deaths (mortality rate, 35%) from 27 countries (http://www.who.int/emergencies/mers-cov/en/) (Figure 1). The evolutionary origins of MERS-CoV are still uncertain. Epidemiologic surveys showed that most of the dromedary camels in Middle East region are serological or MERS-CoV viral nucleic acid positive (9). Several MERS-CoV viruses have also been isolated from these camels indicating that they could be an intermediate host for MERS-CoV (10). In addition, MERS-CoV shares strong sequence similarities with bat CoVs, such as HKU4 and HKU5. HKU4 virus even uses the same DPP4 receptor for entry, and two critical mutations could license bat-to-human transmission of MERS-CoV (11). Whether MERS-CoV is originated from bats as SARS-CoV did remains unclear so far. Current evidence indicates that bats are likely to be the original source (12,13), and dromedary camels are considered to be a possible intermediate host for MERS-CoV (10) (Figure 2). MERS-CoV continuously crosses species and transmits from dromedary camels and/or bats to human population, which poses a significant threat to public health. Previous studies indicated that MERS-CoV infection are primarily due to repeated introductions of MERS-CoV from dromedary camels to human, while human to human transmission is limited (14,15). However, the 2015 MERS outbreak in South Korea with dozens of secondary- and tertiary-generation cases raised the concern that MERS-CoV may have adapted to allow a more efficient spread in humans (16).
As for the evolution, CoVs are one of the most rapidly evolving viruses undergoing frequent genetic recombination and mutations. Compared to other RNA viruses, the estimated evolutionary rates in HCoVs are moderate to high (17,18). For MERS-CoV the mutation rate in the complete genome was estimated to be 1.12×103 substitutions per site per year (19), while HCoV-OC43 and HCoV-229E represent an average mutation rate of about 3~6×104 substitutions per site per year (20,21). Deletion mutation also occasionally occurred in the genome of MERS-CoV, especially in the accessory proteins (22). Based on the analysis of MERS-CoV sequences which are available in genbank, diverse MERS-CoV strains are circulating in dromedaries and human to date, including Clade A and Clade B. Clade B can be further divided into five lineages (lineage 1–5 or group 1–5) (16,19). Evidence of genetic recombinant has also been found in MERS-CoV (16), as well as in other HCoVs, such as HCoV-OC43 (23), HCoV-NL63 (21), HCoV-HKU1 (24), SARS-CoV (25,26). Several groups also reported multiple recombinant MERS-CoV prevalent in dromedary camels and humans (26). The MERS-CoV outbreak in South Korea in 2015 also revealed a probable recombinant event between two different parental Clade B viruses (16). Genetic recombination in MERS-CoV indicates that frequent co-infections with different lineages of MERS-CoV could occur in camels and humans (26). In summary, mutation and deletion in accessory proteins and genetic recombination play a major role in the evolution of MERS-CoV.
Virology and structure
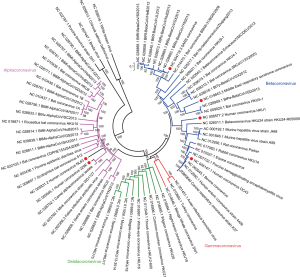
Coronaviruses are the largest positive strand RNA viruses (26–32 kb) that are about 125 nm in diameter (27). Four coronavirus genera have been identified including alpha- (group 1), beta- (group 2), gamma- (group 3) and deltacoronavirus (group 4) genera (28). HCoVs are among the alphacoronavirus (HCoV-NL63 and HCoV-229E) and betacoronavirus (HCoV-HKU1, HCoV-OC43, SARS-CoV and MERS-CoV) (Figure 3). Four HCoVs, including HCoV-NL63, HCoV-OC43, HCoV-229E and HCoV-HKU1, are circulating globally in human population and primarily contribute to 10–20% common cold (3,29), while MERS-CoV and SARS-CoV are the two major causes of severe pneumonia in humans (30,31), other four HCoVs sporadically causing severe pneumonia were also reported (32-35). In 2002–2004 pandemic, SARS-CoV infected over 8,000 patients and resulted in more than 800 deaths (36). MERS-CoV is the etiological agent responsible for the ongoing MERS pandemic in Middle East region. No specific vaccine and drug have so far been licensed for human use (37).
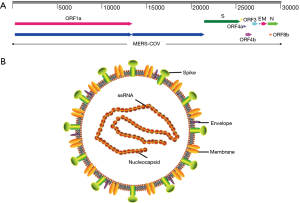
MERS-CoV genome contains a 5'terminal cap structure along with poly (A) tails at the 3'end, the replicase gene encoding the non-structural protein makes up approximately two-third of the genome at the 5' end of genome, which contains 16 non-structural proteins(nsp1-16). Four structural protein, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) protein, and five accessory proteins (ORF3, ORF4a, ORF4b, ORF5 and ORF8) make up about 10kb at the 3' end of genome (Figure 4A). In summary, MERS-CoV genome is typically arranged in the order of 5’terminal-ORF1a-ORF1b-S-E-M-N-3’terminal, accessory proteins are interspersed along the structural genes. The viral membrane contains S, E and M protein, and spike protein plays vital roles in viral entry. MERS-CoV attaches human host-cell receptor dipeptidyl peptidase 4 (hDPP4, CD26) (38) via receptor binding domain (RBD) of spike protein (39). M and E proteins play important role in viral assembly, N protein is required for RNA synthesis (Figure 4B) (28).
Animal models
Robust animal models for MERS infection are urgently needed to elucidate MERS pathogenesis and develop antiviral drugs and vaccines. However, small laboratory animals that generally used for emerging virus studies, such as mice (40,41), ferrets (42), guinea pig (43) and hamster (44) are not susceptible to MERS-CoV infection since their homologous DPP4 molecules do not fit as the receptors for MERS-CoV entry. No effective viral replications were detected in these animals after challenged with high dose of MERS-CoV (45). Upon intratracheally (IT) and intranasally (IN) inoculation with MERS-CoV, New Zealand white rabbits remained free of clinical sing of disease (46), whereas viral RNA was detected in the respiratory tract, and moderate necrosis was observed in nasal turbinates. Dromedary camels, as a reservoir of MERS-CoV, developed mild upper respiratory infections after MERS-CoV infection (47). Alpacas, a close relative within the Camelidae family, secreted live virus after oronasal infection and remained asymptomatic without showing any upper or lower respiratory tract diseases (48,49). In addition, due to their cost and relatively larger size, these animal models are not suitable for high-throughput research for MERS.
Nonhuman primates
Nonhuman primates are useful models for pathogenesis studies and vaccine evaluations for a lot of human infectious diseases. MERS-CoV caused transient lower respiratory tract infection in rhesus macaques. Clinical signs were observed by 1-day post infection (dpi) and resolved as early as 4dpi. No fatal cases observed in rhesus macaque model, the infection is not lethal to the rhesus macaque. This model can be used for studying the pathogenesis of mild MERS-CoV infection in humans (50). The use of common marmosets is still controversial. While a study showed that the infection could developed a progressive severe pneumonia (51), other groups found that common marmosets only developed mild to moderate nonlethal respiratory diseases through intratracheal infection with MERS-CoV (52).
Mice
Wild-type rodents are not susceptible to MERS-CoV infection (41). However, researchers have developed several models that rendered mice susceptible to MERS-CoV infection (53-55). In 2014, the first mouse model for MERS infection was generated (53). In this study, mice were transduced intranasally with recombinant adenovirus 5 encoding hDPP4 molecule. This model supports MERS-CoV replication in the lungs, and mice developed signs of interstitial pneumonia, including inflammatory cells infiltration, alveolar thickening and mild edema (53). This model enables research to use mice for MERS study although there are some limitation, such as uncontrolled level of the hdpp4 expression and their tissue distribution. In 2015, the hDPP4 transgenic mice have been developed (54). These mice could be efficiently infected by MERS-CoV. However, global hDPP4 expression leaded to multiple organ damage (54), resulting in the death of the animals probably due to the lethal brain infection, as observed in ACE2 transgenic mice infected with SARS-CoV (56). Most recently, several MERS mouse models have been generated by replacing mouse DPP4 gene with homologous human DPP4 gene (55,57). Li and colleagues developed human DPP4 knockin (KI) mice, where mouse DPP4 gene fragments had been replaced by homologous human DPP4 fragments responsible for receptor binding. Further they serially passaged WT MERS-CoV in the respiratory tract of these mice for 30 times, resulting a mouse adapted MERS-CoV strain (MERSma). MERSma contained 13–22 mutations that caused significant weight loss and mortality in human DPP4-KI mice (55), this model is so far the best mouse model for MERS.
Immunopathogenesis
The lack of human autopsy data as well as good animal models hindered our understanding on the immunity and pathogenesis of MERS-CoV infection. Based on our knowledge from the studies of SARS, several factors could be involved in MERS pathogenesis, including viral and host factors, interferon induction, dysregulation of cytokines and adaptive immune responses (58,59).
Virus and host interactions
SARS and MERS are both severe pneumonias caused by novel CoVs and they shared some similarities in clinical and laboratory features (58). For instances, the elderly and immunocompromised individuals are more susceptible to both SARS-CoV and MERS-CoV infections (60), However, the average comorbidity rate are much higher in MERS patients than that of SARS patients (76% vs. 10–30%) (61,62). The mortality rate of MERS so far is about 35% which is much higher than that of SARS (~10%) (7). Although some of the mild MERS patient were not readily identified in Middle East region (63), it might also partially due to the difference immunopathogenesis of these two viruses. Unlike SARS-CoV, MERS-CoV could efficiently infect human dendritic cells (64) and macrophages (65) in vitro which would help the virus to dysregulate the immune system. MERS-CoV also has the ability to infect T cells through their highly expression of CD26, leading to T cell apoptosis (66), which might potentially disrupt anti-viral T cell responses. As published previously, the clearance of MERS-CoV and SARS-CoV both required virus-specific T cell responses (67,68).
Some host factors might also be involved in MERS-CoV infection. As the increasing expression level of prostaglandinD2 (PGD2) in aged lungs impaired respiratory DC migration from lung to the draining lymph nodes, which in turn diminished the anti-viral CD8 T cell responses and resulted in increased mortality following SARS-CoV infection (69). Whether any host factors are involved in MERS-CoV infection and increase mortality rate in patients with comorbidity remain unknown.
Interferon antagonism
It is generally accepted that innate immune response is essential for the control of coronavirus infection, and it also determines the extent of initial virus replication and immune response activation (70). MERS-CoV replication is highly sensitive to type I interferon (IFN-I) treatment in cell culture suggesting that IFN-I treatment could be a possible therapeutic approach in clinical practice (71). The combination of IFN-α2b or IFN-β1a with ribavirin was effective in reducing MERS-CoV replication in vitro and improves outcome in MERS-CoV-infected rhesus macaques (72,73). It is well known that CoVs have developed several strategies to evade the innate immune response. In SARS-CoV infection, SARS-CoV accessory proteins ORF3b and ORF6 decreased IFN-expression (74), and ORF6 inhibited nuclear translocation of STAT1 which is the key molecule governing the expression of interferon-stimulated genes (ISG) with antiviral activity (75-77). Similarly, MERS-CoV structural and accessory proteins, including M, ORF4a, ORF4b, and ORF5 had all been proved that could antagonize IFN-I signaling and inhibit ISG productions (78,79).
Antibody and T cell responses
Convalescent serum from MERS and SARS patients could accelerate virus clearance (80,81). Neutralizing antibodies generated in vitro or by vaccination could efficiently prevent the secondary infection with the same strain of CoVs in animal models (82). However, antibody response in patients previously infected with SARS-CoV and MERS-CoV tend to be short lived (68,83). On the other hand, T cell response often target highly conserved internal proteins and are long lived. SARS-CoV-specific memory T cells but not B cells could be detected6 years after infection in SARS survivors (84). A recent study showed that CD8 T cell response could be detected in patients with undetectable neutralizing antibody in convalescent MERS patients (63). Immunodominant epitopes recognized by T cells in MERS-CoV infected mice were found in structural protein S, M, N (53,85). These MERS-CoV-specific CD8 T cells efficiently lysed the target cells in cytotoxicity assays (53). It also had been shown previously, that adoptive transfer of SARS-CoV-specific CD4 or CD8 T cells reduced virus titers in the infected mouse lungs (85).
Vaccine and antiviral drug developments
Vaccine
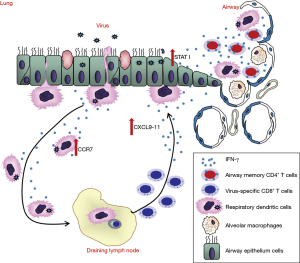
There were still no vaccines and effective antiviral therapeutics against MERS-CoV infection (86). The spike protein of MERS-CoV, which is responsible for MERS-CoV entry is considered as a key target for vaccine development against MERS-CoV infection (87). Multiple vaccine candidates targeting S protein were developed, including DNA vaccines (88,89), subunit vaccines (90,91) and recombinant vector vaccines (92,93). DNA vaccine expressing MERS-CoV S1 gene induced antigen-specific humoral and cellular immune responses in mice (88). In addition, RBD fragment induced the highest-titer IgG antibodies in mice compared with other region of S protein (90). Recombinant vectors (modified vaccinia Ankara and adenovirus vector) expressing MERS-CoV S glycoprotein showed immunogenic in mice (94). Attenuated live vaccine also has been shown to be protective; however the worries of its inadequate attenuation hindered its application (95). The protective role of virus specific CD4T cells is less studied, especially in respiratory CoV infections (63). Most recently, Zhao and colleagues showed that airway memory CD4 T cells were generated after intranasal vaccination with CoV N protein and mediated protection in following CoV challenge (68). These cells could upregulated anti-viral innate response at very early stage of infection and promoted CD8 T cell response by accelerating rDC migration and CD8 T cell mobilization (68). More importantly, they also found that these CD4 T cells targeting a conserved epitope within N protein cross reacted with several other CoVs, indicating that induction of airway memory CD4 T cells should be considered as a component of any universal human coronavirus vaccine (68) (Figure 5). The combination of memory CD4 T cells with those able to elicit strong neutralizing antibodies and memory CD8 T cells could provide better protection against MERS-CoV as well as other respiratory CoV infections.
mAb and antiviral drugs
Neutralizing monoclonal antibodies bind to MERS-CoV spike protein and prevent virus-entry and following membrane fusion, therefore inhibit viral replication and reduce clinical diseases in animal models and humans (86). Neutralizing human monoclonal antibody (mAb) can be used for prophylactic and post-exposure treatments. Several potent mAbs have been developed from MERS infected patient (96), by humanizing mouse mAb against MERS RBD (97) or by screening human antibody phage library (82). All of these mAbs targeted RBD region of spike protein (98). To avoid viral escape mutants, combination of at least two monoclonal antibodies targeting different regions of spike protein had been prove to be more effective (86).
In addition, several antiviral drugs had been developed, including peptide fusion inhibitor targeting heptad-repeat region (HR) of S2 (99), small molecular entry inhibitor targeting S (100), IFN-β, IFN-γ and ribavirin (101). Recently, GS-5734, small-molecule monophosphoramidate, prodrug of an adenosine analog which targeted RdRp, had been found to inhibit MERS-CoV and SARS-CoV replication in multiple in vitro systems (102).
Summary
With the MERS-CoV circulating in the dromedary camels in the Middle East region, efforts regarding epidemic surveillance, phylogenetic analysis, vaccine and antiviral drug developments were still needed to response to the global public health threat posed by this virus. Unlike SARS-CoV, the virus and host interaction as well as pathogenesis and immune responses after MERS-CoV infection in animals and humans are less investigated. Understanding these basic informations will not only enhance emerging CoV research but also will aid our public health preparedness against MERS-CoV.
Acknowledgements
Funding: This work was supported by funds from National Natural Science Foundation of China (NSFC; grants 81772191 and 81702047), State Key Laboratory of Respiratory Disease, 2015 Thousand Talents Plan Award of China and the Municipal Healthcare Joint-Innovation Major Project of Guangzhou (201604020011).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Almeida JD, Tyrrell D. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J Gen Virol 1967;1:175-8. [Crossref] [PubMed]
- Zaki AM, Van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814-20. [Crossref] [PubMed]
- Pyrc K, Berkhout B, van der Hoek L. The novel human coronaviruses NL63 and HKU1. J Virol 2007;81:3051-7. [Crossref] [PubMed]
- Peiris J, Guan Y, Yuen K. Severe acute respiratory syndrome. Nature medicine 2004;10:S88. [Crossref] [PubMed]
- Myint SH. Human coronavirus infections. The Coronaviridae. Springer; 1995:389-401.
- Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 2003;362:1353-8. [Crossref] [PubMed]
- de Groot RJ, Baker SC, Baric RS, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 2013;87:7790-2. [Crossref] [PubMed]
- Kim KH, Tandi TE, Choi JW, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak in South Korea, 2015: epidemiology, characteristics and public health implications. J Hosp Infect 2017;95:207-13. [Crossref] [PubMed]
- Briese T, Mishra N, Jain K, et al. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio 2014;5:e01146-14. [Crossref] [PubMed]
- Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 2014;370:2499-505. [Crossref] [PubMed]
- Yang Y, Liu C, Du L, et al. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol 2015;89:9119-23. [Crossref] [PubMed]
- Ithete NL, Stoffberg S, Corman VM, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis 2013;19:1697. [Crossref] [PubMed]
- Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 2014;16:328-37. [Crossref] [PubMed]
- Sikkema RS, Farag EA, Himatt S, et al. Risk Factors for Primary Middle East Respiratory Syndrome Coronavirus Infection in Camel Workers in Qatar During 2013–2014: A Case-Control Study. J Infect Dis 2017;215:1702-5. [Crossref] [PubMed]
- Hemida M, Elmoslemany A, Al-Hizab F, et al. Dromedary camels and the transmission of Middle East respiratory syndrome coronavirus (MERS-CoV). Transbound Emerg Dis 2017;64:344-53. [Crossref] [PubMed]
- Wang Y, Liu D, Shi W, et al. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. MBio 2015;6:e01280-15. [Crossref] [PubMed]
- Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24:490-502. [Crossref] [PubMed]
- Jenkins GM, Rambaut A, Pybus OG, et al. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol 2002;54:156-65. [Crossref] [PubMed]
- Cotten M, Watson SJ, Zumla AI, et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio 2014;5:e01062-13. [Crossref] [PubMed]
- Ren L, Zhang Y, Li J, et al. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci Rep 2015;5:11451. [Crossref] [PubMed]
- Pyrc K, Dijkman R, Deng L, et al. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol 2006;364:964-73. [Crossref] [PubMed]
- Liu DX, Fung TS, Chong KK-L, et al. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res 2014;109:97-109. [Crossref] [PubMed]
- Lau SK, Lee P, Tsang AK, et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol 2011;85:11325-37. [Crossref] [PubMed]
- Woo PC, Lau SK, Huang Y, et al. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch Virol 2005;150:2299-311. [Crossref] [PubMed]
- Stanhope MJ, Brown JR, Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect Genet Evol 2004;4:15-9. [Crossref] [PubMed]
- Sabir JS, Lam TT-Y, Ahmed MM, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016;351:81-4. [Crossref] [PubMed]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 2015;1282:1-23. [Crossref] [PubMed]
- Monto AS, Cowling BJ, Peiris JM. Coronaviruses. Viral infections of humans. Springer; 2014. p. 199-223.
- Gerna G, Campanini G, Rovida F, et al. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol 2006;78:938-49. [Crossref] [PubMed]
- Chan JF, Lau SK, To KK, et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 2015;28:465-522. [Crossref] [PubMed]
- Graham RL, Donaldson EF, Baric RS. A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 2013;11:836-48. [Crossref] [PubMed]
- Vabret A, Mourez T, Dina J, et al. Human coronavirus NL63, France. Emerg Infect Dis 2005;11:1225. [Crossref] [PubMed]
- Vabret A, Dina J, Gouarin S, et al. Detection of the new human coronavirus HKU1: a report of 6 cases. Clin Infect Dis 2006;42:634-9. [Crossref] [PubMed]
- Pene F, Merlat A, Vabret A, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis 2003;37:929-32. [Crossref] [PubMed]
- Simon A, Völz S, Fleischhack G, et al. Human coronavirus OC43 pneumonia in a pediatric cancer patient with down syndrome and acute lymphoblastic leukemia. J Pediatr Hematol Oncol 2007;29:432-4. [Crossref] [PubMed]
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-66. [Crossref] [PubMed]
- Jiang L, Wang N, Zuo T, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med 2014;6:234ra59-ra59.
- Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013;495:251-4. [Crossref] [PubMed]
- Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013;500:227-31. [Crossref] [PubMed]
- Cockrell AS, Peck KM, Yount BL, et al. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J Virol 2014;88:5195-9. [Crossref] [PubMed]
- Coleman CM, Matthews KL, Goicochea L, et al. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol 2014;95:408-12. [Crossref] [PubMed]
- Raj VS, Smits SL, Provacia LB, et al. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J Virol 2014;88:1834-8. [Crossref] [PubMed]
- de Wit E, Feldmann F, Horne E, et al. Domestic Pig Unlikely Reservoir for MERS-CoV. Emerg Infect Dis 2017;23:985. [Crossref] [PubMed]
- de Wit E, Prescott J, Baseler L, et al. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PloS One 2013;8. [Crossref] [PubMed]
- Peck KM, Scobey T, Swanstrom J, et al. Permissivity of Dipeptidyl Peptidase 4 Orthologs to Middle East Respiratory Syndrome Coronavirus Is Governed by Glycosylation and Other Complex Determinants. J Virol 2017;91:e00534-17. [Crossref] [PubMed]
- Haagmans BL, van den Brand JM, Provacia LB, et al. Asymptomatic Middle East respiratory syndrome coronavirus infection in rabbits. J Virol 2015;89:6131-5. [Crossref] [PubMed]
- Haagmans BL, van den Brand JM, Raj VS, et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science 2016;351:77-81. [Crossref] [PubMed]
- Adney DR, Bielefeldt-Ohmann H, Hartwig AE, et al. Infection, replication, and transmission of Middle East respiratory syndrome coronavirus in alpacas. Emerg Infect Dis 2016;22:1031. [Crossref] [PubMed]
- Crameri G, Durr PA, Klein R, et al. Experimental infection and response to rechallenge of alpacas with Middle East respiratory syndrome coronavirus. Emerg Infect Dis 2016;22:1071. [Crossref] [PubMed]
- Yao Y, Bao L, Deng W, et al. An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J Infect Dis 2014;209:236-42. [Crossref] [PubMed]
- Falzarano D, de Wit E, Feldmann F, et al. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog 2014;10. [Crossref] [PubMed]
- Johnson RF, Via LE, Kumar MR, et al. Intratracheal exposure of common marmosets to MERS-CoV Jordan-n3/2012 or MERS-CoV EMC/2012 isolates does not result in lethal disease. Virology 2015;485:422-30. [Crossref] [PubMed]
- Zhao J, Li K, Wohlford-Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci 2014;111:4970-5. [Crossref] [PubMed]
- Zhao G, Jiang Y, Qiu H, et al. Multi-organ damage in human dipeptidyl peptidase 4 transgenic mice infected with Middle East respiratory syndrome-coronavirus. PloS One 2015;10. [Crossref] [PubMed]
- Li K, Wohlford-Lenane CL, Channappanavar R, et al. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci 2017;114:E3119-E28. [Crossref] [PubMed]
- Tseng C-TK, Huang C, Newman P, et al. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human Angiotensin-converting enzyme 2 virus receptor. J Virol 2007;81:1162-73. [Crossref] [PubMed]
- Cockrell AS, Yount BL, Scobey T, et al. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2016;2:16226. [Crossref] [PubMed]
- Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015;386:995-1007. [Crossref] [PubMed]
- Arabi YM, Balkhy HH, Hayden FG, et al. Middle East respiratory syndrome. N Engl J Med 2017;376:584-94. [Crossref] [PubMed]
- Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med 2014;160:389-97. [Crossref] [PubMed]
- Fowler RA, Lapinsky SE, Hallett D, et al. Critically ill patients with severe acute respiratory syndrome. JAMA 2003;290:367-73. [Crossref] [PubMed]
- de Wit E, van Doremalen N, Falzarano D, et al. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 2016. [Crossref] [PubMed]
- Zhao J, Alshukairi AN, Baharoon SA, et al. Recovery from the Middle East respiratory syndrome is associated with antibody and T cell responses. Sci Immunol 2017.2. [PubMed]
- Chu H, Zhou J, Wong BH-Y, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology 2014;454-455:197-205. [Crossref] [PubMed]
- Zhou J, Chu H, Li C, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis 2014;209:1331-42. [Crossref] [PubMed]
- Chu H, Zhou J, Wong BH-Y, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis 2016;213:904-14. [Crossref] [PubMed]
- Channappanavar R, Zhao J, Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol Res 2014;59:118-28. [Crossref] [PubMed]
- Zhao J, Zhao J, Mangalam AK, et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 2016;44:1379-91. [Crossref] [PubMed]
- Zhao J, Zhao J, Legge K, et al. Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest 2011;121:4921. [Crossref] [PubMed]
- Zuniga EI, Macal M, Lewis GM, et al. Innate and Adaptive Immune Regulation During Chronic Viral Infections. Annu Rev Virol 2015;2:573-97. [Crossref] [PubMed]
- de Wilde AH, Raj VS, Oudshoorn D, et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J Gen Virol 2013;94:1749-60. [Crossref] [PubMed]
- Falzarano D, De Wit E, Rasmussen AL, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nature Med 2013;19:1313. [Crossref] [PubMed]
- Shalhoub S, Farahat F, Al-Jiffri A, et al. IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J Antimicrob Chemother 2015;70:2129-32. [Crossref] [PubMed]
- Spiegel M, Pichlmair A, Martinez-Sobrido L, et al. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 2005;79:2079-86. [Crossref] [PubMed]
- Zhao J, Falcon A, Zhou H, et al. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J Virol 2009;83:2368-73. [Crossref] [PubMed]
- Zhou H, Ferraro D, Zhao J, et al. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J Virol 2010;84:3542-51. [Crossref] [PubMed]
- Frieman M, Yount B, Heise M, et al. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 2007;81:9812-24. [Crossref] [PubMed]
- Rabouw HH, Langereis MA, Knaap RC, et al. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 2016;12. [Crossref] [PubMed]
- Yang Y, Zhang L, Geng H, et al. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 2013;4:951. [Crossref] [PubMed]
- Chan KH, Chan JFW, Tse H, et al. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect 2013;67:130-40. [Crossref] [PubMed]
- Simmons G, Reeves JD, Rennekamp AJ, et al. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci 2004;101:4240-5. [Crossref] [PubMed]
- Tang XC, Agnihothram SS, Jiao Y, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci 2014;111:E2018-E26. [Crossref] [PubMed]
- Hilgenfeld R, Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res 2013;100:286-95. [Crossref] [PubMed]
- Tang F, Quan Y, Xin ZT, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol 2011;186:7264-8. [Crossref] [PubMed]
- Zhao J, Zhao J, Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol 2010;84:9318-25. [Crossref] [PubMed]
- Du L, Yang Y, Zhou Y, et al. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets 2017;21:131-43. [Crossref] [PubMed]
- Pascal KE, Coleman CM, Mujica AO, et al. Pre-and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci 2015;112:8738-43. [Crossref] [PubMed]
- Chi H, Zheng X, Wang X, et al. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine 2017;35:2069-75. [Crossref] [PubMed]
- Al-Amri SS, Abbas AT, Siddiq LA, et al. Immunogenicity of Candidate MERS-CoV DNA Vaccines Based on the Spike Protein. Sci Rep 2017;7:44875. [Crossref] [PubMed]
- Tai W, Wang Y, Fett CA, et al. Recombinant receptor-binding domains of multiple Middle East respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J Virol 2016;91:e01651-16. [Crossref] [PubMed]
- Wang L, Shi W, Joyce MG, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun 2015;6:7712. [Crossref] [PubMed]
- Gilbert SC, Warimwe GM. Rapid development of vaccines against emerging pathogens: The replication-deficient simian adenovirus platform technology. Vaccine 2017;35:4461-4. [Crossref] [PubMed]
- Kim E, Okada K, Kenniston T, et al. Immunogenicity of an adenoviral-based Middle East Respiratory Syndrome coronavirus vaccine in BALB/c mice. Vaccine 2014;32:5975-82. [Crossref] [PubMed]
- Song F. Characterization of recombinant Modified Vaccinia virus Ankara for delivery of Middle East Respiratory Syndrome Coronavirus spike protein antigens: lmu; 2014. Available online: https://edoc.ub.uni-muenchen.de/17246/1/Song_Fei.pdf
- Modjarrad K. MERS-CoV vaccine candidates in development: The current landscape. Vaccine 2016;34:2982-7. [Crossref] [PubMed]
- Corti D, Zhao J, Pedotti M, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci 2015;112:10473-8. [Crossref] [PubMed]
- Chen Y, Lu S, Jia H, et al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg Microbes Infect 2017;6. [Crossref] [PubMed]
- Li Y, Wan Y, Liu P, et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res 2015;25:1237. [Crossref] [PubMed]
- Lu L, Liu Q, Zhu Y, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun 2014;5:3067. [Crossref] [PubMed]
- Liu Q, Xia S, Sun Z, et al. Testing of Middle East respiratory syndrome coronavirus replication inhibitors for the ability to block viral entry. Antimicrob Agents Chemother 2015;59:742-4. [Crossref] [PubMed]
- Chan JF-W, Yao Y, Yeung M-L, et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis 2015;212:1904-13. [Crossref] [PubMed]
- Sheahan TP, Sims AC, Graham RL, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med 2017.9. [PubMed]