Viral infection in community acquired pneumonia patients with fever: a prospective observational study
Introduction
Community acquired pneumonia (CAP) is a common disease, with a reported mortality rate of 7.3% in Asia, based on a large prospective multicentre study (1). In Germany, the mortality rate of adult CAP was 8.6%, outpatients and inpatients had the mortality rate of 0.8% and 12.2% respectively (2). The plague of influenza A (H1N1) in 2009 caused serious viral pneumonia with high mortality rates in a large part of the world. In addition, severe hypoxemic acute respiratory failure and acute respiratory distress syndrome (ARDS) were strongly associated with influenza A (H1N1) in 2009 (3,4). The influenza outbreak was so severe that extracorporeal membrane oxygenation (ECMO) was used to decrease death (5,6).
A multiplex polymerase chain reaction (mPCR) method was a quick and effective diagnostic strategy (7,8) used by many clinical researchers and led to higher positive rates of pathogens associated with pneumonia. Compared with traditional cell culture, antigen assays, and serology, mPCR has improved accuracy and efficiency for the viral diagnosis (9). Many studies have focused on the incidence and aetiology of CAP patients seen at emergency departments and the intensive care unit (ICU) (10,11), and analysed the clinical characteristics and outcomes of viral CAP. To our knowledge, this is a prospective study on the pathogens of CAP inpatients with a fever using an established enrolment system. Most CAP patients suffer from fever, which is typical for many diseases, but there are few studies on virus-associated CAP patients with fever in non-epidemic seasons. In this study, we examined the incidence of viral pneumonia in CAP inpatients with fever and the associated risk factors or indicative laboratory indexes for viral infection and outcomes, such as death during hospitalization
Methods
Patients
This prospective study was conducted through our enrolment system: (I) feverish adult (body temperature ≥37.5 °C, aged ≥18 years inpatients in Shanghai pulmonary hospital from January 2015 to October 2016 were enrolled in this system; (II) chest X-rays or CT scans were conducted as soon as all the fever patients were admitted to the hospital. A new infiltrate on the chest radiograph of these fever patients was diagnosed as CAP, according to the criteria of the British Thoracic Society (12); (III) nasopharyngeal swab and sputum samples were collected from enrolled CAP patients after admission to the hospital; (IV) mPCR kits were used to detect viruses in both nasopharyngeal swabs and sputum samples. During the study period, samples of swab and sputum were done for mPCR within 72 hours after collection; (V) clinical data were systematically collected by physicians and analysed using statistical software. The CURB-65 score consists of 5 points: confusion, BUN >7 mmol/L, respiratory rate >30 breaths/min, systolic blood pressure <90 mmHg and/or diastolic blood pressure ≤60 mmHg, and age ≥65 yrs (13). This project was approved by the ethical committee of Shanghai Pulmonary Hospital.
Samples and detection method
After admission to the hospital, we collected all the nasopharyngeal swab and sputum samples from the patients for viral detection. Samples of swab and sputum were done for mPCR within 72 hours after collection. Viral nucleic acids were purified from each nasopharyngeal swab or sputum specimen using the Axyprep Body Fluid Viral DNA/RNA Miniprep Kit, following the manufacturer’s instructions. The resultant DNA/RNA was eluted with 60 µL elution buffer and stored at −80 °C until testing. Ten kinds of viral DNA/RNA [influenza A and B, adenovirus (ADV), respiratory syncytial virus (RSV) A and B, picornavirus, parainfluenza virus (PIV), coronavirus, human metapneumovirus (HMPV), and bocavirus] were detected using the one step primeScript RT-PCR kit (Perfect Real-time, DRR064; Takara) according to the manufacturer’s instructions. The RT-PCR assay conditions were: 42 °C for 5 min, 95 °C for 10 s, 40 cycles of 95 °C for 3 s and 60 °C for 25 s with the CFX96 real-time PCR detection system (Bio-Rad). Blood, bronchoalveolar lavage fluid, and sputum samples were collected to obtain general information on the bacteria, which were cultured by the laboratory clinicians of our hospital.
Microbiological diagnosis
Virus infection confirmed by positive RT-PCR on nasopharyngeal swab or sputum specimen. Bacterial infection was identified by one of the following sample culture positive: (I) bronchoalveolar lavage fluid (threshold 104 CFU/mL); (II) blood culture; (III) sputum culture (considering >25 neutrophils and <10 epithelial cells as a valid specimen, threshold 105 CFU/mL).
Statistical analysis
Clinical data, including demographic variables such as age, gender, clinical symptoms, underlying diseases and complications, severity score assessed by CURB65, laboratory indexes, and outcomes were collected by our physicians. Statistical analyses were performed in SPSS (version 24.0). Dichotomous variables were analysed with chi-squared or Fisher’s exact test. The Mann-Whitney U test or Kruskal-Wallis method was used to analyse numerical data. Risk factors associated with severe situations, death during hospitalization, analysed by multiple logistic regression. Logistic regression analysis was also used to examine the risk factors associated with influenza A viral infection. A P value ≤0.05 was considered statistically significant.
Results
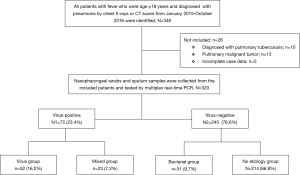
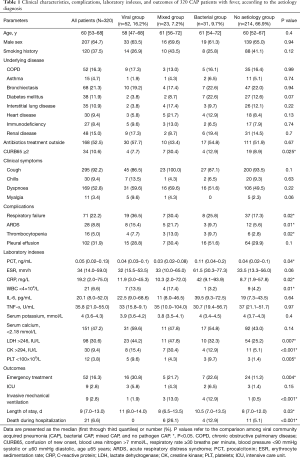
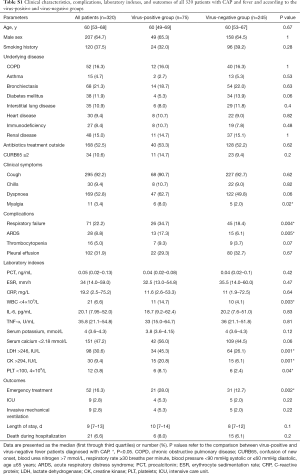
The 320 patients enrolled in our study were divided into four categories (viral group, mixed group, bacterial group, and no pathogen group) (Figure 1) according to the causative agent of pneumonia, and we also classified all the patients into two sub-groups (viral-positive and viral-negative) based on viral infection or not. The characteristics of clinical symptoms, complications, and outcomes according to the aetiology diagnosis are shown in Tables 1,S1. In this study, 64.7% were male with a median age of 60 years (IQR, 53–68 years). There was no statistically significant difference among the four categories of underlying diseases. Approximately one third of patients in the mixed-group had a CURB65 ≥2 score that was higher than the other three groups. The median hospitalization span was 9 days (IQR, 7.0–13.0 days). When examining outcomes according to the pathogens (Table 1), the mixed-group had the highest incidences of death in the hospital. Laboratory indexes are shown in Table 1. As to the two subgroups (viral-positive and viral-negative), there were 71 (22.2%) and 28 (8.8%) patients with respiratory failure and ARDS, respectively, and the virus-positive group had a larger population for these two complications than the virus-negative group (Table S1). Virus-positive patients had higher serum lactic dehydrogenase and creatine kinase (CK) compared with virus-negative patients (Table S1).
Full table
Full table
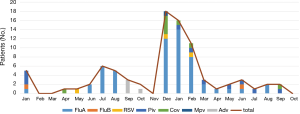
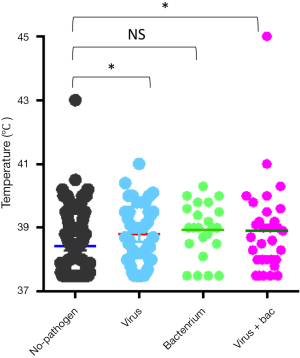
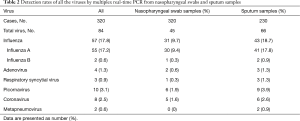
The most common virus detected in CAP patients with fever was influenza A, followed by picornavirus. The epidemic seasons are autumn and winter and viral detection peaks were in December and February (Figure 2). The distribution of the highest body temperature during the whole course, according to the pathogen group, is shown in Figure S1. The viral-group and mixed group were more likely to have a higher fever (T >39 °C) than the no pathogen group. The mPCR positive results for nasopharyngeal swab samples and sputum samples are shown in Table 2. The causes of bacterial co-infections in the mixed group are shown in Table S2. Streptococcus pneumoniae followed by haemophilus influenzae were the most common bacteria in co-infections.
Full table

Full table
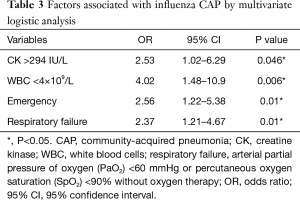
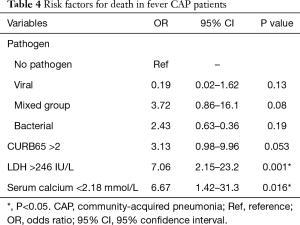
Influenza virus was critical for viral pneumonia in our study. In addition, CK >294 IU/L, WBC <4×109/L, emergency admission, and respiratory failure were independent factors associated with influenza infection in multivariate logistic regression analysis (Table 3). In this study, we also analysed two risk factors for outcomes of CAP patients with fever, i.e., death. Viral CAP and mixed CAP were not independent factors for death. In addition, lactate dehydrogenase (LDH) >246 IU/L, and serum calcium <2.18 mmol were associated with death (Table 4).
Full table
Full table
Discussion
The purpose of this study was to analyse the aetiology, incidence, clinical characteristics, and outcomes of CAP patients with fever during non-pandemics in one pulmonary hospital. This study revealed there was a viral aetiology in 75 (23.4%) of 320 fever patients diagnosed with CAP. This is in line with other studies, where the rates of viral infection in CAP patients varied from 23–56% (10,11,14,15). The differences in aetiologies of CAP patients between studies is likely due to the different test methods, the diversity of the aetiologies detected, and the distinctions of different regions and populations. During the observational period in our study, the viral infection peak months were from December to February. The predominant virus was influenza A followed by picornavirus, and streptococcus was the most common bacteria in viral co-infection cases, which is consistent with the results from other studies (16). de Roux et al. reported 65% of mixed pneumococcal infections were associated with a respiratory virus in hospitalized patients, mainly the influenza virus (17). Another study suggested that pneumococcus is critical for the development of pneumonia associated with influenza virus A, RSV, PIV, and ADV (18).
In our study, most viral-positive patients, with or without bacterial co-infection, had high temperatures. This finding is in contrast to the common opinion that high temperatures frequently occur in patients with bacterial or mixed infections. This discrepancy may be attributed to the difficulty in identifying a bacterial infection because of the use of antibiotics before testing. Although, this result also indicated that fever cannot be an empirical indicator for antibiotic treatment because a bacterial infection may have been misdiagnosed before any diagnostic tests.
In our study, nasopharyngeal swabs and sputum samples were collected for mPCR. Only 230 patients had sputum samples, which had high detection rates for viruses, suggesting there is a lower viral load in nasopharyngeal samples from CAP patients (19). Furthermore, this also suggested that sensitivity of viral pathogen identification could be enhanced by simultaneous examination both NPS and sputum samples rather than only NPS samples (20). Influenza virus is the most common virus in most reports; although, the presence of myalgia and neutropenia were associated with influenza pneumonia in the study by Jennings et al. (21). Furthermore, we identified that having influenza pneumonia increased the odds of emergency room visits and respiratory failure; while, previous reports revealed that patients admitted with influenza who have pneumonia are more likely to be admitted to the ICU or die (22,23). The more accurate the detection of influenza infection, the more correct the use of antivirals will be and unnecessary antibiotics use will be reduced among patients. In our study, high levels of CK were also associated with influenza pneumonia; therefore, CK may be a predictive marker in influenza infection. The combined use of biologic markers and viral testing may correctly identify patients and safely limit antibiotic exposure.
Due to pre-hospitalization antibiotic usage in the majority of subjects, the rates of bacterial and viral-bacterial detection may be underestimated in our study. Interesting, in our research, only one case had bacteremia which was diagnosed with positive blood culture, which was lower than other studies (10,24,25). Approximately one third of subjects with viral pneumonia had bacterial co-infection. This rate was lower than found in other studies (10), but we observed mild-to-moderate patients; while, other studies focused on severe CAP patients, which may have led to differences. In our study, we found that the co-infection group had the highest death rate (21.6%) compared with the other three groups, but co-infection was not an independent risk factor for death during hospitalization. The lower mortality in our mild-to-moderate subjects should be re-inspected in a much larger study group. The mixed group was also identified as an independent risk factor for complicated courses (death and invasive mechanical ventilation) in ICU samples (10). This trend may be explained by the finding that influenza virus (predominant virus in co-infection with bacterium) neuraminidase could contribute to excess mortality in bacterial pneumonia (26). Possible explanations for mixed CAP progressing into severe cases include epithelial damage, changes in airway function, up-regulation of receptors, and changes in the innate immune response (27). Previous study reports that patients with co-infection were much more likely to have a pneumonia severity index (PSI) score of IV or V and a longer length of stay (10). The influenza outbreak was so severe that ECMO was used to decrease death. Influenza plays a dominant role in co-infection, which was so severe that needed effective means to decrease death. The Influenza patients diagnosed with severity score CURB 65 and detection method mPCR can be admitted to ICU sooner and start treatment like ECMO and Tamiflu sooner with increased survival (5,6).
There were some limitations in our study. The single centre design restricted the observed populations. We used cultures instead of mPCR for bacterial detection. The mPCR method can detect dead as well as viable bacteria (14); therefore, the culture method would have lower positive rates of bacterial and co-infection. Moreover, we underestimated the role of atypical pathogens such as C. pneumonia and M. pneumonia that would manifest fever in most cases. The low mortality rate could impair the accuracy of the independent risk factors in the multiple logistic regression analysis.
In conclusion, we found strong variations in the virus distribution in non-pandemic years with relatively high viral rates in the CAP population with fever. It is likely that ARDS and respiratory failure are associated with viral infection, even though there are no definite predictors for causative virus in CAP. The role of co-infection in CAP is unclear. It was not an independent risk factor for death but LDH >246 IU/L [odds ratio (OR) =7.06, 95% confidence interval (CI): 2.15–23.2, P=0.001], and serum calcium <2.18 mmol/L (OR =6.67, 95% CI: 1.42–31.3, P=0.016) were associated with death. The implications of positive testing can offer the physicians accurate information with early treatment of influenza, and then to prevent progression to CAP and respiratory failure. More studies on the treatment of viral CAP are necessary because effective antiviral therapy may prevent the development of CAP and improve the outcomes.
Acknowledgements
Funding: This work was supported by the program from National Natural Science Foundation of China (NSFC 81670006), National key research and development program (2016YFC0903801), the Key Program of the Shanghai Hospital Development Center (SHDC12014104, 16CR3036A) and Shanghai Leading Talent Program (2016036).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the ethical committee of Shanghai Pulmonary Hospital (K17-113) and written informed consent was obtained from all patients.
References
- Song JH, Oh WS, Kang CI, et al. Epidemiology and clinical outcomes of community-acquired pneumonia in adult patients in Asian countries: a prospective study by the Asian network for surveillance of resistant pathogens. Int J Antimicrob Agents 2008;31:107-14. [Crossref] [PubMed]
- Welte T, Kohnlein T. Global and local epidemiology of community-acquired pneumonia: the experience of the CAPNETZ Network. Semin Respir Crit Care Med 2009;30:127-35. [Crossref] [PubMed]
- Kumar A, Zarychanski R, Pinto R, et al. Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA 2009;302:1872-9. [Crossref] [PubMed]
- Estenssoro E, Rios FG, Apezteguia C, et al. Pandemic 2009 influenza A in Argentina: a study of 337 patients on mechanical ventilation. Am J Respir Crit Care Med 2010;182:41-8. [Crossref] [PubMed]
- Rozencwajg S, Brechot N, Schmidt M, et al. Co-infection with influenza-associated acute respiratory distress syndrome requiring extracorporeal membrane oxygenation. Int J Antimicrob Agents 2018;51:427-33. [Crossref] [PubMed]
- Huang L, Zhang W, Yang Y, et al. Application of extracorporeal membrane oxygenation in patients with severe acute respiratory distress syndrome induced by avian influenza A (H7N9) viral pneumonia: national data from the Chinese multicentre collaboration. BMC Infect Dis 2018;18:23. [Crossref] [PubMed]
- Poritz MA, Blaschke AJ, Byington CL, et al. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 2011;6. [Crossref] [PubMed]
- Stockton J, Ellis JS, Saville M, et al. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol 1998;36:2990-5. [PubMed]
- van Elden LJR, van Kraaij MGJ, Nijhuis M, et al. Polymerase chain reaction is more sensitive than viral culture and antigen testing for the detection of respiratory viruses in adults with hematological cancer and pneumonia. Clin Infect Dis 2002;34:177-83. [Crossref] [PubMed]
- Voiriot G, Visseaux B, Cohen J, et al. Viral-bacterial coinfection affects the presentation and alters the prognosis of severe community-acquired pneumonia. Crit Care 2016;20:375. [Crossref] [PubMed]
- Das D, Le Floch H, Houhou N, et al. Viruses detected by systematic multiplex polymerase chain reaction in adults with suspected community-acquired pneumonia attending emergency departments in France. Clin Microbiol Infect 2015;21:608.e1-8. [Crossref] [PubMed]
- Lim WS, Baudouin SV, George RC, et al. BTS guidelines for the management of community acquired pneumonia in adults: update 2009. Thorax 2009;64 Suppl 3:iii1-55. [Crossref] [PubMed]
- Lim WS, van der Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax 2003;58:377-82. [Crossref] [PubMed]
- Gadsby NJ, Russell CD, McHugh MP, et al. Comprehensive Molecular Testing for Respiratory Pathogens in Community-Acquired Pneumonia. Clin Infect Dis 2016;62:817-23. [Crossref] [PubMed]
- Angeles Marcos M, Camps M, Pumarola T, et al. The role of viruses in the aetiology of community-acquired pneumonia in adults. Antivir Ther 2006;11:351-9. [PubMed]
- Huijskens EG, van Erkel AJ, Palmen FM, et al. Viral and bacterial aetiology of community-acquired pneumonia in adults. Influenza Other Respir Viruses 2013;7:567-73. [Crossref] [PubMed]
- de Roux A, Ewig S, Garcia E, et al. Mixed community-acquired pneumonia in hospitalised patients. Eur Respir J 2006;27:795-800. [Crossref] [PubMed]
- Madhi SA, Klugman KP. Vaccine Trialist G. A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med 2004;10:811-3. [Crossref] [PubMed]
- Casiano-Colon AE, Hulbert BB, Mayer TK, et al. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J Clin Virol 2003;28:169-74. [Crossref] [PubMed]
- Yoshii Y, Shimizu K, Morozumi M, et al. Identification of pathogens by comprehensive real-time PCR versus conventional methods in community-acquired pneumonia in Japanese adults. Infect Dis (Lond) 2016;48:782-8. [Crossref] [PubMed]
- Jennings LC, Anderson TP, Beynon KA, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax 2008;63:42-8. [Crossref] [PubMed]
- Louie JK, Acosta M, Winter K, et al. Factors associated with death or hospitalization due to pandemic 2009 influenza A(H1N1) infection in California. JAMA 2009;302:1896-902. [Crossref] [PubMed]
- Lee N, Chan PKS, Lui GCY, et al. Complications and Outcomes of Pandemic 2009 Influenza A (H1N1) Virus Infection in Hospitalized Adults: How Do They Differ From Those in Seasonal Influenza? J Infect Dis 2011;203:1739-47. [Crossref] [PubMed]
- Laterre PF, Garber G, Levy H, et al. Severe community-acquired pneumonia as a cause of severe sepsis: Data from the PROWESS study. Crit Care Med 2005;33:952-61. [Crossref] [PubMed]
- Voiriot G, Dury S, Parrot A, et al. Nonsteroidal Antiinflammatory Drugs May Affect the Presentation and Course of Community-Acquired Pneumonia. Chest 2011;139:387-94. [Crossref] [PubMed]
- Peltola VT, Murti KG, McCullers JA. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis 2005;192:249-57. [Crossref] [PubMed]
- McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 2006;19:571-82. [Crossref] [PubMed]


