Urinary circulating DNA profiling in non-small cell lung cancer patients following treatment shows prognostic potential
Introduction
Lung cancer is a major cause of cancer related mortalities globally (1), and is often detected in the advance stage. Much focus has been dedicated to early detection (2,3) where the primary tumor is largely localized. Early disease is effectively treated via surgery or in combination with radiotherapy (3). For patients with non-metastatic lung cancer, disease recurrence is a cause for concern (4,5). Following surgical resection, and possibly combined radio- and chemo-therapy approaches, routine clinical surveillance relies heavily on longitudinal radiographic scans during follow-up visits (4). Microscopic tumors may be missed and limits the usefulness of imaging scans in early detection (6). In addition, radiography does not detect genetic aberrations that arise during disease development and are critical in treatment decisions (7).
Alternative methods using serological measurements have been proposed for lung cancer management. The sample recovery is less invasive, which makes these alternative strategies attractive and suitable for serial long-term monitoring. Circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) from plasma have been most successful (8,9) to address various challenges in non-small cell lung cancer (NSCLC) management (10). For instance, lung tumor tissues may acquire a series of somatic mutations during disease progression. Certain mutations such as EGFR T790M confers drug resistance and affects overall survival of patients (11,12). These had been demonstrated to be effectively captured from CTCs (13) and ctDNA (14). Previous studies had also shown excellent clinical correlation to advance stage NSCLC patients (15-17). In other studies, detection of plasma DNA was used for treatment monitoring (18) and disease prognosis (19). The key challenge for clinical adoption is the low frequency of ctDNA and CTC in plasma that requires pre-enrichment and relatively large volume of blood extraction. Measurements from urine specimens present an attractive substitute and prior studies had demonstrated presence of nucleic acid in urine that originated from the body’s circulatory system (20,21). Urine specimens can be collected in relatively large volumes and easy to extract unlike tissue biopsies. In prior studies that used urinary DNA in NSCLC, clinical utility was demonstrated for late stage NSCLC patients. It was also observed that there was a strong disease correlation and the marker performed well as a predictor for overall survival (22,23). There is a growing body of evidences that demonstrates the clear usefulness of urinary DNA (24,25).
In the current study, the use of urinary DNA to identify relapse cases in NSCLC is explored. In addition, the study aims to investigate the relationship of urinary DNA in minimal residual disease (MRD) for patients who underwent curative surgery. Addressing pre- and post-treatment, variations in urinary DNA concentrations were examined.
Methods
Study design and NSCLC patient characteristics
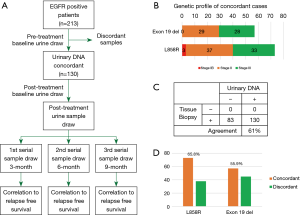
The samples analyzed for this study were collected prospectively from an initial pool of 213 patients. Participants provided informed consent and procedures were approved by the Institutional Review Board (No. NB235/02134). In all cases, patients enlisted had good quality matched tumor samples and were EGFR positive with either L858R point mutation or Exon 19 deletions. These patients had localized lung cancer and underwent surgical resection. A summary of their clinico-pathological parameters is presented in Table 1. Patients with concordant EGFR profiles detected from urinary DNA with matched tumor tissues at baseline were subsequently measured at specific time intervals. Following baseline measurement, the first post-treatment sampling was taken within 3 weeks of surgery for each patient. Subsequent samplings were consecutively recovered at 3-month intervals for a total duration of 9 months.

Full table
Urine sample processing
All urine samples taken at different time intervals were analyzed for mutant EGFR using digital droplet PCR (ddPCR). The pre-analytic processes involved for urine sample preparations comprised of two centrifugation steps to remove debris and contaminants. To ensure consistency in sample collection, only the first morning urine was taken. Urine specimen containers were prefilled with EDTA (0.5 M, pH 8.0, Sigma Aldrich, China) and approximately 30 mL of urine was collected during each measurement. Samples were processed within 3 hours after collection to minimize degradation. To prevent contaminants, raw urine samples were spun at 5,000 g in a centrifuge (Eppendorf 5417, Eppendorf AG, USA) for 15 minutes at 4 °C. The supernatant was carefully transferred to a clean 50 mL centrifuge tube. A repeat centrifugation step using the same setting was used; 16 mL of processed urine was recovered for DNA extraction.
Urinary DNA extraction and EGFR profiling
For DNA extraction from urine, we utilized the Qiagen QIAmp Circulating Nucleic Acid Kit (Qiagen Inc., USA) following the manufacturer’s protocols. DNA purification was achieved with multiple DNA immobilization steps using spin columns processed on the micro-centrifuge. Purified DNA was eluted in 10 µL TE buffer supplied by the manufacturer; 1 µL of sample was used on the NANODROP 2000 photospectrometer (Thermofisher Scientific, USA) to quantify the amount of nucleic acid obtained. Samples with low quantity or quality had a repeated urine draw the following day. For DNA profiling, the specimens were batched and probed using the QX200 ddPCR system (Bio-Rad, USA). Primers and probes for both EGFR L858R and Exon 19 deletions for the PCR reactions were available as a commercial kit from the PrimePCR ddPCR mutation assay (Bio-Rad, USA). The reaction setup followed closely recommended guidelines provided by the manufacturer. All components were thawed at room temperature and mixed by vortexing to ensure homogeneity. A master mix was then prepared depending on the sample size for the batch. Negative and positive controls were added in each run for confirmation of a successful test. Each reaction mix comprised of the following: 1× concentration of ddPCR Supermix for Probes (10 µL); 1× concentration of target primers (1 µL); 1× concentration of reference primers/probes (1 µL); restriction enzyme (1 µL); DNA template diluted in nuclease free water (7 µL). Once the reaction mixtures were ready, 70 µL of droplet generation oil were added into each reaction and placed in the QX200 droplet generator (Bio-Rad, USA). After droplet generation, the sample plate was transferred for thermal cycling. Thermocycling conditions comprised of 1 cycle at 95 °C for 10 minutes; 40 cycles at 95 °C for 30 s, 55 °C for 1 minute; 1 cycle at 98 °C for 10 minutes for final extension. Each reaction plate was analyzed using the QX200 droplet reader and readout from the QuantaSoft software (Bio-Rad, USA).
Statistical analysis
Concordance between matched tumor tissue and urinary DNA specimens was ascertained at baseline pre-treatment. Comparison of urinary DNA concentrations post-treatment was performed using a Student t-test. A repeat measure analysis of variance was conducted to address the variability during longitudinal measurements. All variables defined are represented as mean and standard deviation unless otherwise specified. Disease-free survival was conducted using the Kaplan-Meier method with log-rank test to estimate the P value and Cox regression for the hazard ratios (HR). For multivariate analysis, a logistics regression model was used. Calculations were performed using the PRISM software (Version 6.0, GraphPad Inc., USA).
Results
NSCLC patient genetic profiling and baseline concordance
In the current study, NSCLC patients were prospectively recruited and urine specimens were batched purified and processed. All 213 participants with localized NSCLC were proven by biopsy and radiographic imaging. The study addressed patients with variations in mutant EGFR DNA profiles and thus our inclusion criteria included only patients with EGFR mutations tested using tumor tissue. EGFR L858R and exon 19 deletions are two most common forms of mutations in NSCLC, and were examined in our study. Among all participants, 6 were stage IB, 104 stage II and 103 stage III disease based on the AJCC TNM classification. The mean age of patients was 53 years old and majority were of adenocarcinoma histology. Table 1 highlights the background information of the study group.
Among the 213 patients, 130 were sub-selected for further monitoring based on the baseline urinary DNA results. Figure 1A illustrates the study design and the sample collection intervals. Not all patients received adjuvant therapy during the entire monitoring period. The treatment cycles were determined by their respective attending physicians and were based on a comprehensive examination from radiographic imaging and other related tests. A common trait among these 130 patients were detectable EGFR mutations identified using ddPCR in their urine specimens. A breakdown of the patients’ disease profiles is shown in Figure 1B, with 3 in stage IB and majority in stage II and III (n=127). Based on the current results, the concordance rate of NSCLC patients with detectable urinary DNA in stage IB, II and III were 50%, 63% and 59% respectively. Overall agreement of urine sample testing with tissue biopsy was 61% (Figure 1C). Figure 1D shows the separation based on mutational profiles for patient results concordant with tissue profiling. Samples with consistent results from urinary DNA testing and tumor tissue profiling with EGFR L858R and exon 19 deletions were 65.8% and 55.9% respectively. To correlate the changes in urinary DNA quantity to disease relapse and possibly the presence of MRD, these 130 patients were serially monitored and their urine tested at regular intervals.
Quantity of mutant DNA at different measurement points
A total of 733 samples were processed during the entire study. The variation in pre- and post-treatment mutant DNA concentrations for each corresponding patient is shown in Figure 2A. Mean detected concentration for pre-treatment NSCLC patients was 220±237 copies/mL. During post-treatment, we observed a significant decline in mean concentration of 74% from 130 patients (59±103 copies/mL, P<0.001, Student’s t-test). Of these, 61 specimens had undetectable traces of mutant DNA. All patients experienced a drop in mutant DNA content, with the minimum decline registered at 1.96%.
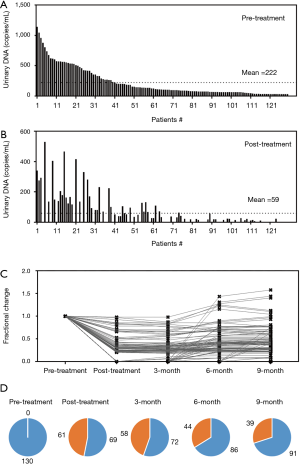
To explore the serial urinary DNA concentration variability within the patient group, we normalized the results with baseline concentrations. The trend lines are depicted in Figure 2B. Each line represents the result from 1 of the 130 patients that were examined post-treatment. We observed an increase in detectable urinary DNA among the 130 patients following the first post-treatment results from 53.1% to 55.4%. For the subsequent second and third measurements, the detectable sample size increases from 66.2% and 70.0% respectively. Within the patient group with undetectable mutations in the urinary DNA (Figure 2C), it was interesting that for these NSCLC patients with localized disease, persistent detection of mutant DNA within the urine specimen exist. We also observed significant variability for the study cohort and a repeat measure ANOVA of the data at each time point showed significant differences in post-treatment results.
Correlation to disease-free survival
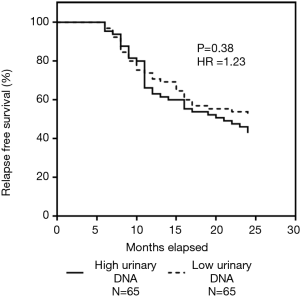
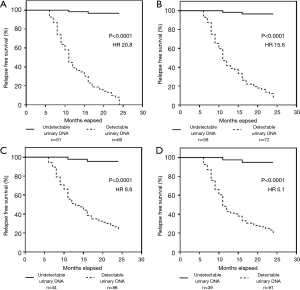
In subsequent analysis, we explored whether the detection of urinary DNA for EGFR was associated with outcome. Within a follow up period of 24 months, we observed 68 patients with disease relapse, accounting for 52.3% of the total 130 serially monitored patients. Our results strongly indicated that the presence of detectable urinary DNA is clearly linked to patients with disease recurrence as shown in Figure 3. In Figure 3A, the study cohort was divided based on detectable urinary DNA immediately post-treatment. The results showed a significant outcome with patients having undetectable urinary DNA to have better disease-free survival (P<0.0001). The HR as determined using the Cox regression model was 20.8 (95% CI: 12.3–35.4). Similar results were obtained at the other time point measurements as shown in Figure 3B,C,D. Results at 3-, 6- and 9-month indicated significant differences in the disease relapse outcome based on detectable urinary DNA from patients. The corresponding HR were 15.6 (95% CI: 9.3–26.2), 6.6 (95% CI: 4.0–10.8) and 5.1 (95% CI: 3.3–7.9) respectively. As a comparison, we analyzed the results of pre-treatment patients as shown in Figure S1. HR was determined to be 1.23 (P=0.38).
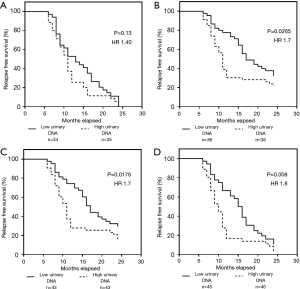
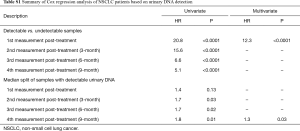
In a further analysis, we examined whether the patients with lower urinary DNA detected had possibly better disease outcome. For the study cohort, we divided equally into two groups based on the median detected concentration of urinary DNA. Interestingly, we observed insignificant differences in outcome at the measurement period immediately post-treatment (Figure 4A) with HR of 1.40 (95% CI: 0.9–2.4, P=0.13). Similar analysis was performed at the other time points as shown in Figure 4B,C,D. Based on this preliminary data, these results were significant and HR values was observed to increase at later time points. The results indicated better predictions at later time points with increased HR values. At the 9-month, the HR value was 1.8 (95% CI: 1.2–3.4). The intermediate measurements points at 3- and 6-month saw HR of 1.7 (95% CI: 1.1–2.9) and 1.7 (95% CI: 1.1–3.1) respectively. This possibly indicate that the concentration of mutant EGFR content in urinary DNA could potentially be used to better address the monitoring needs of these patients. A summary of the results is provided in Table S1. A multivariate analysis was conducted taking into consideration all other factors. The results showed significant distinction in the first post treatment measurement for detectable and undetectable urinary DNA samples. This was similarly true for the subset of detectable urinary DNA patient group for the last measurement. Adjusted for all other covariates, the last measurement at 9-month showed significant differences with the control group.
Full table
Discussion
Our results suggest that urine testing can be a promising approach to understand challenges in NSCLC with localized disease. This can be a good complementary assay for existing clinical imaging modalities. In this current study, we examined EGFR positive patients to better address residual disease after curative intent surgical procedures and its association to disease relapse. The clear benefits of this approach are being noninvasive and routine regular testing will allow better predictions to disease recurrence. In baseline measurements of 213 NSCLC cases, we noted good clinical correlations for EGFR mutations using urinary DNA, which reflected a strong clinical relevance to the disease. Subsequent serial measurements of urine samples at post-treatment phase demonstrated good clinical utility to stratify patients with low risk of disease relapse, particularly those with undetectable mutant DNA.
In baseline reference measurements, we observed a significant prevalence in positive mutation detection in NSCLC patients with localized disease. This highlights one major limitation in current imaging techniques that is unable to actively probe disease changes. The positive identification of EGFR mutations in urine could also imply the presence of micro-metastases currently undetectable by radiographic imaging. Especially for NSCLC patients with persistently positive EGFR mutations at post-treatment phase, this indicated possible residual disease that may warrant further treatment or more intense monitoring for disease recurrence. We noted our tumor tissue agreement with urinary DNA is consistent with several recent studies using plasma DNA as the sampling fluid (26,27). Fernandez-Cuesta et al. in early stage patients found approximately 49% of small cell lung cancer (SCLC) cases with concordant TP53 mutations (27). This gene is important as it encodes for a tumor suppressor protein p53. In our case for NSCLC, EGFR is a prevalent mutation and strongly linked to the efficacy of tyrosine kinase inhibitor (TKI) therapy and detecting this mutation is critical for first line therapeutic selections. It is interesting that we observed a large dispersion in the detected copies and the rate of concordance is evenly spread across both EGFR mutations. This highlights the heterogeneity in the disease and possibly linked to the existing conditions of pre-treated patients. In a related study, Diehl et al. noted that the cancer dynamics of colorectal cancer patients is closely represented by circulating mutant DNA and related to tumor volume (28). We hypothesize that the levels of urinary DNA detected within our patients could similarly indicate tumor burden associations. A more detailed analysis will need to be conducted to ascertain the relationship. Going beyond detection of urinary DNA, our observed variability during pre- and post-treatment may possibly indicate treatment efficacy and residual disease after therapy.
To address this hypothesis, we analyzed the results of four post monitoring time points in an attempt to chart out the disease dynamics. For cases of detectable mutant DNA with respect to pre-treatment conditions, varying amount of changes were observed. In a small number of cases, we noticed that the concentration of mutant urinary DNA exceeded pre-treatment values. Among the detectable mutant DNA cases post-treatment, we noticed a slight increase possibly as a result of disease progression. Indeed, as we performed the KM estimator on different time points, patients with undetectable mutant DNA had better relapse free survival in all cases. This is consistent with a recent study by Chaudhuri et al. (29) where pre- and post-treatment plasma DNA were analyzed. NSCLC patients had better outcomes in cases with undetectable plasma DNA. Interestingly more than half of their study cohort with early disease had plasma DNA profiles with sensitizing genetic mutations to TKIs. This can aid in treatment of early disease when these mutations are detected. Consequently, our patient groups with EGFR L858R or exon 19 deletions detected at post-treatment stage could possibly benefit from further secondary treatment. This will require further verifications in a large scale prospective clinical intervention study. To better understand the role of detecting urinary DNA in these patients, we addressed the group with detectable urinary DNA content and performing the KM estimator based on concentration levels. Other than the reading immediately post-treatment, all three other time points had statistical significant outcome. The results were conclusive to show that early disease patients with lower urinary DNA concentrations had better disease-free outcome. This points to the need for more intense monitoring of patients with higher urinary DNA concentrations as they may be at higher risk of disease recurrence. Overall, this shows the clinical usefulness of urinary DNA for post-treatment patient risk stratification and disease surveillance.
Conclusions
In conclusion, we demonstrated that urinary DNA is a promising biomarker for early NSCLC. Within a controlled group of EGFR mutation positive patients, we observed a relatively large variability in urinary DNA concentrations that could be used to indicate residual disease. Continual monitoring of patients is critical even after surgical procedures and urinary DNA can aid to predict disease recurrence. The study forms the basis of future prospective trials in clinical interventions.
Acknowledgements
Funding: This study was supported by a research grant provided by the Ningbo Provincial Clinical Trial Specialty Grant (2013-88).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by institutional/regional/national ethics committee/ethics board of Ningbo No. 2 Hospital (No. NB235/02134) and written informed consent was obtained from all patients.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Henschke CI, McCauley DI, Yankelevitz DF, et al. Early Lung Cancer Action Project: overall design and findings from baseline screening. Lancet 1999;354:99-105. [Crossref] [PubMed]
- Hirsch FR, Franklin WA, Gazdar AF, et al. Early detection of lung cancer: clinical perspectives of recent advances in biology and radiology. Clin Cancer Res 2001;7:5-22. [PubMed]
- Crino L, Weder W, van Meerbeeck J, et al. Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2010;21 Suppl 5:v103-15. [Crossref] [PubMed]
- Fruh M, De Ruysscher D, Popat S, et al. Small-cell lung cancer (SCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24 Suppl 6:vi99-105. [Crossref] [PubMed]
- Bruzzi JF, Munden RF. PET/CT imaging of lung cancer. J Thorac Imaging 2006;21:123-36. [Crossref] [PubMed]
- Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012;264:387-96. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539-46. [PubMed]
- Ettinger DS, Wood DE, Akerley W, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. J Natl Compr Canc Netw 2016;14:255-64. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Yeo T, Tan SJ, Lim CL, et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci Rep 2016;6:22076. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6. [Crossref] [PubMed]
- Xu S, Lou F, Wu Y, et al. Circulating tumor DNA identified by targeted sequencing in advanced-stage non-small cell lung cancer patients. Cancer Lett 2016;370:324-31. [Crossref] [PubMed]
- Jiang T, Ren S, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer 2015;90:128-34. [Crossref] [PubMed]
- Qiu M, Wang J, Xu Y, et al. Circulating tumor DNA is effective for the detection of EGFR mutation in non–small cell lung cancer: a meta-ANALYSIS. Cancer Epidemiol Biomarkers Prev 2015;24:206-12. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446. [Crossref] [PubMed]
- Zhong XY, Hahn D, Troeger C, et al. Cell-free DNA in urine: a marker for kidney graft rejection, but not for prenatal diagnosis? Ann N Y Acad Sci 2001;945:250-7. [Crossref] [PubMed]
- Tong YK, Lo YM. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta 2006;363:187-96. [Crossref] [PubMed]
- Li F, Huang J, Ji D, et al. Utility of urinary circulating tumor DNA for EGFR mutation detection in different stages of non-small cell lung cancer patients. Clinical and Translational Oncology 2017;19:1283-91. [Crossref] [PubMed]
- Chen S, Zhao J, Cui L, et al. Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clinical and Translational Oncology 2017;19:332-40. [Crossref] [PubMed]
- Franovic A, Raymond VM, Erlander MG, et al. Urine test for EGFR analysis in patients with non-small cell lung cancer. Journal of thoracic disease 2017;9:S1323. [Crossref] [PubMed]
- Singh AP, Li S, Cheng H. Circulating DNA in EGFR-mutated lung cancer. Ann Transl Med 2017;5:379. [Crossref] [PubMed]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Fernandez-Cuesta L, Perdomo S, Avogbe PH, et al. Identification of Circulating Tumor DNA for the Early Detection of Small-cell Lung Cancer. EBioMedicine 2016;10:117-23. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Chaudhuri AA, Chabon JJ, Lovejoy AF, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 2017;7:1394-403. [Crossref] [PubMed]