High CCR4 expression in the tumor microenvironment is a poor prognostic indicator in lung adenocarcinoma
Introduction
Lung cancer is the most common cause of cancer-related death globally, accounting for over a million deaths per year (1). Cancer immunotherapy has elicited great interest over the past several years due to the development of immune checkpoint blockade therapy achieving durable long-term responses of refractory malignancies, including lung cancers in a minority of patients (2-5). Immune responses within the tumor microenvironment are now acknowledged to be important factors determining tumor progression and aggressiveness, as well as responsiveness to immune-modulating agents. Types and densities of tumor-infiltrating immune cells, and their expression of immune genes and cytokines have been widely explored for their application as prognostic markers in lung cancer (6-8).
Chemokines are a group of structurally-related small soluble mediators which can induce directional migration of different lymphocyte subsets, and are also known to play important roles in the immune response to cancer (9). CC chemokine receptor 4 (CCR4) is a member of the CC chemokine receptor family to which bind the TARC/CCL17 and MDC/CCL22 chemokines that are produced by tumor cells and tumor-infiltrating macrophages. High levels of CCR4 expression are characteristic of certain CD4+ T-cells including regulatory T cells (Tregs), Th2 and Th17 cells, but not Th1 cells (9-14). It is of note that unlike Th1 cells, Tregs and Th17 CD4+ T cells are widely acknowledged as elements creating immune suppressive microenvironments (15-17).
Recent studies have shown that the anti-CCR4 monoclonal antibody (mAb) mogamulizumab selectively depletes effector Tregs from the tumor microenvironment and may augment immune responses in several solid cancers, including lung cancer (11-13). If the infiltration of CCR4+ lymphocytes is a poor prognostic factor in lung cancer, these patients may be suitable candidates for anti-CCR4 mAb monotherapy or in combination with other immunomodulating agents. In fact, elevated expression of CCR4 in tumors is generally associated with poor prognosis in several cancers, including gastric cancer, breast cancer, and renal cell carcinoma (18-20). However, little is known about the roles of CCR4+ lymphocytes and their prognostic impact in lung cancer, especially in lung adenocarcinoma.
In the present study, we first used established public gene expression databases as a discovery dataset and determined that higher CCR4 gene expression in the tumor may be a poor prognostic factor in lung adenocarcinoma. We then constructed tissue microarrays (TMAs) to validate the public database analysis at the level of protein expression and localization, and to further evaluate the prognostic impact of CCR4 expression by strictly controlling for confounding factors.
Methods
Analysis of CCR4 gene expression and prognosis using public databases
To analyze the association of CCR4 expression and prognosis, a large pooled database of the results of Affymetrix microarray gene expression assessments was used (KM-plotter, http://www.kmplot.com/lung) (21-23). Gene expression and clinical data were extracted and integrated from the Cancer Biomedical Informatics Grid, the Gene Expression Omnibus, and The Cancer Genome Atlas. The follow-up threshold was set at 10 years and patients surviving >10 years were censored. Biased arrays were excluded. These were defined as those having two or more of the following parameters out of the 95% range of all arrays: percentage of present calls, background, raw Q value, presence of bioB-/C-/D-spikes, GAPDH and ACTB ratio 3 to 5. The list of Affymetrix probe ID selected for analysis is shown in Table S1. We initially used the database version 2013, although during the present study, this was updated from version 2013 to version 2015. Thus, herein we present results using database version 2015 which contains a larger amount of patient data.
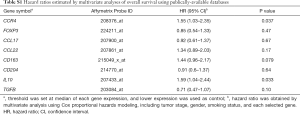
Full table
Patient selection for the TMA assay
Clinicopathological records of 677 lung cancer patients who underwent surgery in our Department from August 2004 to December 2011 were retrospectively reviewed. To control for confounding factors, inclusion criteria were set as follows: diagnosis by pathology as primary lung adenocarcinoma, complete resection by lobectomy (either by open thoracotomy or VATS), tumor size up to 3 cm (pathological T1 tumor as defined in the 7th edition of TNM staging), and an adequate amount of tissue available. Adenocarcinoma in situ (AIS) and minimally-invasive adenocarcinoma (MIA) were excluded from the study. Those patients who underwent preoperative chemotherapy and/or radiotherapy, and those with a postoperative follow-up <1 month were also excluded. Written informed consent was obtained from each patient and the protocol for this study was approved by the Institutional Review Board of The University of Tokyo Hospital (No. 10334).
Detailed clinicopathological data were collected during hospitalization from the medical records for all patients, including age, gender, smoking history, predominant histological subtype, tumor size, and pathological TNM stage. Follow-up data were collected in October 2016 and also information about recurrence and survival status. Recurrence-free survival (RFS) was defined as the length of time that the patient survived after surgery without recurrence. Overall survival (OS) was measured from surgery to death from any cause, or censored.
Construction of TMA and immunohistochemical staining
All resected tumor samples were retrieved from the archives of the Department of Pathology of The University of Tokyo Hospital to create TMA. Samples were fixed with formalin and embedded in paraffin. Hematoxylin and eosin-stained slides of all the cases were reviewed. Four-micrometer thick sections were subjected to immunohistochemistry on a Ventana Benchmark XT stainer (Ventana Medical Systems, Tucson, AZ, USA). TMA sections were generated according to well-established procedures described previously (24). Briefly, sections were deparaffinized and pretreated with CC1buffer (Roche Diagnostics, Basel, Switzerland) for 60 min, and then incubated with primary antibodies at 37 °C for 32 min. The clones and dilutions of primary antibodies were directed against the following: CD4 (1:30, 1F6, Leica Biosystems, Newcastle, UK), anti-CD8 (1:40, 4B11, Leica Biosystems), and anti-Foxp3 antibody (1:50, PAB12685, Abnova, Taipei). Chromogenic detection was performed with I-VIEW DAB Universal kit (Roche). As for CCR4 staining, Poteligeo test IHC kit (Kyowa Medex, Tokyo, Japan) was used according to the manufacturer’s instructions.
After all slides were digitalized using a digital slide scanner (NanoZoomer, Hamamatsu Photonics, Shizuoka, Japan), immunohistochemical results were evaluated by means of image analysis software (Tissue Studio, Definiens AG, Munich, Germany) (25). Staining thresholds (hematoxylin, DAB density) and morphological criteria (shape, area) were introduced in order to exclude stromal cells and non-cancerous areas. Lymphocytes were considered as CCR4+ TILs based on distinct membrane and cytoplasmic staining, CD4+ and CD8+ TILs were identified by surface staining and for Foxp3 assessment, lymphocytes with clear nuclear staining were considered positive. Numbers of total TILs and positively-stained TILs were calculated and expressed as cell numbers per mm2 of tumor tissue area. The study population was stratified into two groups according to a higher or lower than median value for the respective positively-stained TILs.
Statistical analysis
Statistical analysis of the TMA assay results was performed with SAS v9.4 (SAS Institute, Cary, NC, USA). Online tools were used for analysis of the public database results (KM-plotter) (21). Welch’s t-test was performed to compare continuous variables between groups. The Kaplan-Meier method was used for survival analysis, with significance of differences assessed by the log-rank test. Cox regression analyses were performed to identify prognostic factors; those included in the model were determined by the backward deletion method which sequentially deletes variables with a P value >0.20. Differences were considered statistically significant when the P value was <0.05.
Results
Higher CCR4 gene expression is associated with poor prognosis in the public database analysis
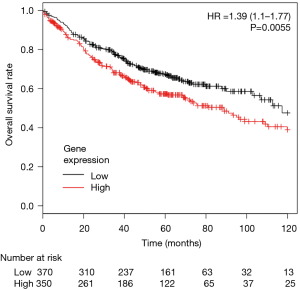
To determine the association between CCR4 expression and prognosis in lung adenocarcinoma patients, we first conducted a study using pooled data from public databases. Of 2,437 lung cancer patients included, 720 adenocarcinoma patients were selected for this analysis. Patients were divided into two groups by the higher or lower than median level of expression of CCR4. Median OS of the lower expression cohort was 117.3 versus 88.7 months for the higher group. This difference was statistically significant [HR =1.39 (95% CI: 1.1–1.77), P=0.0055] (Figure 1).
Next, we conducted multivariate analysis using Cox proportional hazards modeling, including tumor stage, gender, smoking status, and CCR4 gene expression. Multivariate analysis indicated that never having smoked was a good prognostic factor [HR =0.51 (0.3–0.87), P=0.013], whereas tumor stage [HR =2.5 (1.82–3.42), P<0.0001] and high CCR4 gene expression status [HR =1.55 (1.03–2.35), P=0.037] were poor prognostic factors (Table 1).

Full table
Because high CCR4 expression by Tregs has been reported, we also included FOXP3 expression in the analysis, as this is characteristic of Tregs. However, FOXP3 gene expression did not possess prognostic relevance either in uni- or multi-variate analysis [univariate, HR =1.12 (0.88–1.43), P=0.36; multivariate, HR =1.3 (0.86–1.97), P=0.22]. Thus, analyses of publically-available data indicate that high CCR4 gene expression is associated with poor prognosis for lung adenocarcinoma patients, whereas FOXP3 gene expression had no prognostic relevance.
To investigate the underlying etiology between high CCR4 expression and poor prognosis, we additionally analyzed prognostic relevance of gene expressions of CCR4 ligands CCL17 and CCL22 using KM-plotter, although prognostic relevance was not observed [CCL17, HR =0.92 (0.61–1.37), P=0.67; CCL22, HR =1.34 (0.89–2.03), P=0.17] (Table S1). These ligands are produced by M2 macrophages. Thus we further analyzed the gene expression of CD163 and CD204, surface markers of M2 macrophages [CD163, HR =1.44 (0.96–2.17), P=0.079; CD204, HR =0.91 (0.6–1.37), P=0.64]. Patients with high expression of CD163 gene tended to have poor prognosis, although the difference was not statistically significant. We also analyzed prognostic relevance of IL10 and TGFB gene expressions. Tregs produce these inhibitory molecules. IL10 is also produced by M2 macrophages as well as Th2 CD4+ T cells and Tregs. While high expression of TGFB was not associated with poor prognosis [HR =0.71 [0.47–1.07], P=0.10], high IL10 expression was associated with significantly poor prognosis [HR =1.59 (1.04–2.44), P=0.033]. These results indicated that high CCR4 expression may partly reflect immune suppressive microenvironment due to not only Tregs but Th2/M2 responses and thus associated with poor prognosis.
Characteristics of the patients whose tumors were available for the TMA assay
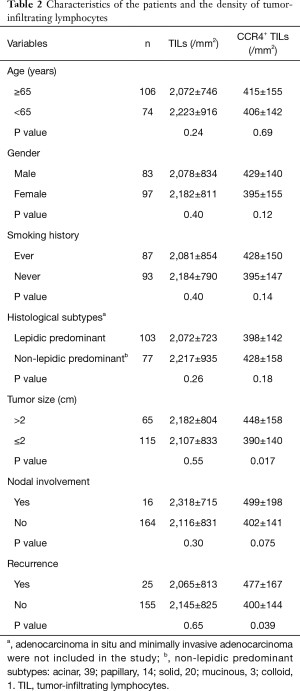
To validate the association between CCR4 expression and prognosis, and to gain further insight into CCR4 expression at the protein level in the tumor microenvironment, we created TMAs of lung adenocarcinoma using locally resected specimens. To control for confounding factors, the cohort was restricted to patients with adenocarcinomas up to 3 cm in size resected by lobectomy with curative intent. AIS and MIA were excluded, and finally 180 tumor samples met these inclusion criteria and were used in this analysis. Patients’ characteristics are shown in Table 2. Eighty three patients were male and 97 female; 87 had a history of cigarette smoking, 66 (75.9%) of which were men. One-hundred and three patients had tumors of a predominantly lepidic histological subtype, with the remaining 77 predominantly non-lepidic (39 acinar, 20 solid, 14 papillary, 3 mucinous, and 1 colloid). Tumor size was >2 cm (pT1b according to the 7th edition of TNM staging) in 65 and ≤2 cm (pT1a) in 115. Pathological N stages were N0 in 164, N1 in 6 and N2 in 10 patients. Median follow-up after surgery was 74 months (range, 1–146 months), during which 25 patients suffered recurrence. Five-year OS of the study cohort was 90.6% and RFS was 82.1%.
Full table
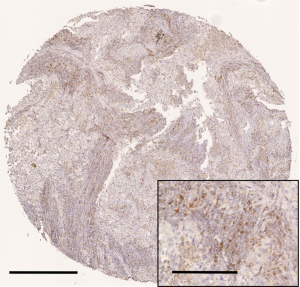
Representative immunostaining for CCR4 is shown in Figure 2. The mean values of CCR4+ TILs and total TIL densities were 411±149/mm2 and 2,134±821/mm2, respectively. CD8-, CD4-, and Foxp3-positive TIL densities were 615±390/mm2, 1,170±758/mm2, and 323±209/mm2, respectively. The total TIL density was not significantly different according to clinicopathological variables, but the density of CCR4+ TILs was significantly higher in tumors >2 cm in size (P=0.017). CCR4+ TIL density was also higher in tumors of patients suffering recurrence (P=0.039). These results thus indicate that CCR4+ lymphocyte infiltration may be associated with tumor progression and hence poor prognosis.
The density of CCR4+ TILs is an independent prognostic factor in lung adenocarcinoma
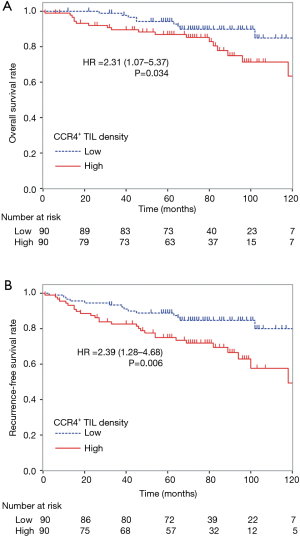
To control for confounding factors and to determine the prognostic impact of CCR4+ TILs, uni- and multivariate analyses including each clinicopathological factor were performed. By univariate analysis, age ≥65 years, male gender, having ever smoked, predominant non-lepidic histological subtype, and nodal involvement were significant factors indicating poor prognosis for both OS and RFS (Table 3). Tumor size >2 cm also associated with poor prognosis for RFS. Although neither CD8+, CD4+, nor Foxp3+ TIL density was significantly associated with the prognosis in the univariate analysis, high CCR4+ TIL density was significantly associated with worse prognosis for both OS and RFS [OS, HR =2.31 (1.07–5.37), P=0.034; RFS, HR =2.39 (1.28–4.68), P=0.006]. The Kaplan-Meier survival curves for these 180 patients with lung adenocarcinomas ≤3 cm stratified according to CCR4+ TIL density are shown in Figure 3.
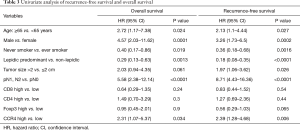
Full table
By multivariate analysis, age ≥65 years [OS, HR =3.78 (1.60–10.44), P=0.0026; RFS, HR =2.85 (1.44–6.07), P=0.0027], male gender [OS, HR =6.17 (2.55–17.03), P<0.0001; RFS, HR =2.91 (1.49–6.02), P=0.0018], nodal involvement [OS, HR =6.19 (2.35–15.92), P<0.0001; RFS, HR =7.30 (3.42–15.31), P<0.0001] and high CCR4+ TIL density [OS, HR =2.24 (1.01–5.34), P=0.049; RFS, HR =2.20 (1.16–4.39), P=0.017] were identified as factors for poor prognosis of both OS and RFS (Table 4). On the other hand, high CD8+ TIL density [OS, HR =0.19 (0.08–0.46), P=0.0001; RFS, HR =0.28 (0.13–0.56), P=0.0002] and predominantly lepidic subtype [RFS, HR =0.25 (0.11–0.54), P=0.0004] were associated with a good prognosis. Consistent with the results of the public database gene expression analysis, Foxp3+ TIL density was not identified as a significant prognostic factor in the TMA assay.
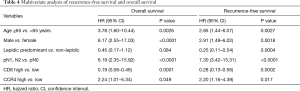
Full table
Taken together, these results strongly suggest that expression of CCR4 at both the gene and protein levels in TILs from lung adenocarcinoma is associated with poor prognosis. Higher expression of CCR4 may therefore mark a tumor microenvironment associated with poor prognosis.
Discussion
In this study, the relevance of the expression of CCR4 in lung adenocarcinoma has been investigated in two different patient cohorts: (I) a discovery cohort from public databases of CCR4 gene expression in tumors, and (II) a validation cohort from surgically-resected patients’ specimens in our institution analyzed for CCR4 protein expression by TILs. The discovery cohort indicated that higher than median CCR4 gene expression was associated with a poorer prognosis, and our validation cohort confirmed that a higher number of CCR4+ TILs was related to poor prognosis. Using multivariate analysis, in both cohorts, higher expression of CCR4 in the tumor microenvironment was indicated as an independent poor prognostic factor. Expression of CCR4 may reflect the existence of a tumor microenvironment related to poor prognosis, and thus evaluation of CCR4 expression may become a novel biomarker for lung adenocarcinoma.
With the ever-increasing monetary cost of cancer treatment including immunotherapy, there is an unmet need to discover suitable biomarkers that will enable selection of those patients most likely to benefit from each immune-modulating agent and to develop novel approaches to convert non-responders into responders to these therapies. Because high CCR4 expression in the tumor microenvironment may be a poor prognostic factor, patients with high CCR4+ lymphocyte infiltration may be suitable candidates for anti-CCR4 antibody treatment. Recent studies have shown promising results using mogamulizumab, an anti-CCR4 antibody which selectively depletes immunosuppressive effector Tregs as well as CCR4+ Th2 and Th17 cells from the tumor microenvironment, and thus may improve immune responses (11-13). Clinical trials of this anti-CCR4 antibody with or without other immune-modulating agents including immune checkpoint blockade therapy are currently underway in Japan (NCT01929486, NCT02476123). However, these clinical studies are not analyzing the influence of the pretreatment level of CCR4 expression. The present study provides evidence suggesting that assessing pretreatment CCR4 expression would be a useful marker for guiding clinical trials with anti-CCR4 mAb and potentially stratifying patients accordingly.
To reveal the underlying etiology between high CCR4 expression and poor prognosis, we additionally analyzed prognostic relevance of gene expressions of CCR4 ligands (CCL17 and CCL22), surface markers of M2 macrophages (CD163 and CD204), and immunosuppressive cytokine associated with Th2/M2 responses and Tregs (IL-10 and TGFB) using public databases. High IL10 expression was associated with significantly poor prognosis. High expression of CD163 gene also tended to have poor prognosis, although the difference was not statistically significant. These results indicated that not only Tregs but Th2 CD4+ T cells and M2 macrophages are involved in the formation of immunosuppressive tumor microenvironment. In this study, CCR4 expression was better than other Th2/M2 markers for elucidating the association between intratumor immunosuppressive condition and poor prognosis. Further investigation is needed to reveal the underlying etiology between high CCR4 expression and poor prognosis.
In the present study, not only CCR4 expression, but age, male gender, predominantly non-lepidic subtype, nodal involvement, and low CD8+ TIL density were also found to be independently associated with poorer prognosis. This result is consistent with recent studies, supporting the generalizability of the present study (26-29), and adding further factors to consider when stratifying patients for tailor-made immunotherapies.
Interestingly, in the present study, no association was shown between FOXP3 expression and prognosis, although by univariate analysis of RFS, higher Foxp3+ TIL density tended to be related to better rather than worse prognosis, but this did not reach statistical significance. These results are not consistent with some of recently published reports on Treg associations (7,30). In addition, correlation between CCR4+ TIL density and Foxp3+ TIL density could not be observed in the present study (Pearson’s R=0.07, P=0.35), although correlation between CD4+ TILs and CCR4+ TILs (R=0.35, P<0.0001), and CD4+ TILs and Foxp3+ TILs (R=0.62, P<0.0001) were statistically significant. These results may be at least partly explained by the analysis of different functional subtypes of Tregs in the different studies because these cells can be classified into three phenotypes: CD45RA+Foxp3low Tregs (naïve Tregs), CD45RA–Foxp3low Tregs (non-effector Tregs) and CD45RA–Foxp3high Tregs (effector Tregs) (31). Among these subtypes, suppression of cancer-immunity is mainly caused by effector Tregs. It has been reported that CCR4 is expressed on immunosuppressive effector Tregs (11,12), while Foxp3 is expressed in all subtypes of Tregs. Considering the subtypes of Tregs and according to staining conditions and cutoff values, we may hypothesize that the number of CCR4+ TILs and Foxp3+ TILs may not necessarily correlate, and the prognostic value of Foxp3 expression may differ. The fact that the prognostic impact of Foxp3 expression is often discussed in the context of the ratio of Foxp3 expression to other markers (i.e., ratio to CD3 or CD4) also suggests the difficulty of using Foxp3 expression as a reliable prognostic biomarker. In fact, for example, in colon cancer, the prognostic impact of Foxp3 Tregs is controversial, although several studies have reported a relationship between better prognosis and high Foxp3 expression, as we tended to see here in adenocarcinoma patients (32,33).
There are several limitations to the present study. First, potential confounding biases could not be completely avoided due to the retrospective nature of both the public database and the TMA analyses. In addition, clinicopathological data and detailed treatment information were not available for all of the patients in the public databases. Also, in the TMA assay, the location of TILs within the tumor was not considered, although this may be more important than their overall numbers. Another limitation of the study is that the cohort used for the TMA assay was limited to pathological T1 tumors (≤3 cm). Although the analysis based on public database included the patients in all stages, associations between CCR4+ lymphocyte infiltration and prognosis in locally-advanced cancer requires further investigation. Importantly, well designed clinical trials are needed for the proof of the concept that anti-CCR4 mAb may benefit the lung adenocarcinoma patients with high CCR4 expression in the tumor microenvironment. The method for assessing CCR4 expression (gene expression assay and/or IHC staining) also needs to be optimized before embarking on clinical trials of anti-CCR4 mAb targeting of tumors based on high CCR4 expression.
Conclusions
Abundant CCR4+ TILs and higher expression of the CCR4 gene were identified as poor prognostic factors in lung adenocarcinoma. Patients with high CCR4 expression in the tumor microenvironment may have a poorer prognosis and thus may be suitable candidates for clinical trials of anti-CCR4 mAb treatment.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Written informed consent was obtained from each patient and the protocol for this study was approved by the Institutional Review Board of The University of Tokyo Hospital (No. 10334).
References
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563-7. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Hellmann MD, Rizvi NA, Goldman JW, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): results of an open-label, phase 1, multicohort study. Lancet Oncol 2017;18:31-41. [Crossref] [PubMed]
- Suzuki K, Kachala SS, Kadota K, et al. Prognostic immune markers in non-small cell lung cancer. Clin Cancer Res 2011;17:5247-56. [Crossref] [PubMed]
- Suzuki K, Kadota K, Sima CS, et al. Clinical impact of immune microenvironment in stage I lung adenocarcinoma: tumor interleukin-12 receptor beta2 (IL-12Rbeta2), IL-7R, and stromal FoxP3/CD3 ratio are independent predictors of recurrence. J Clin Oncol 2013;31:490-8. [Crossref] [PubMed]
- Brambilla E, Le Teuff G, Marguet S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:1223-30. [Crossref] [PubMed]
- Ishida T, Ueda R. CCR4 as a novel molecular target for immunotherapy of cancer. Cancer Sci 2006;97:1139-46. [Crossref] [PubMed]
- Bromley SK, Mempel TR, Luster AD. Orchestrating the orchestrators: chemokines in control of T cell traffic. Nat Immunol 2008;9:970-80. [Crossref] [PubMed]
- Sugiyama D, Nishikawa H, Maeda Y, et al. Anti-CCR4 mAb selectively depletes effector-type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc Natl Acad Sci U S A 2013;110:17945-50. [Crossref] [PubMed]
- Kurose K, Ohue Y, Sato E, et al. Increase in activated Treg in TIL in lung cancer and in vitro depletion of Treg by ADCC using an antihuman CCR4 mAb (KM2760). J Thorac Oncol 2015;10:74-83. [Crossref] [PubMed]
- Kurose K, Ohue Y, Wada H, et al. Phase Ia Study of FoxP3+ CD4 Treg Depletion by Infusion of a Humanized Anti-CCR4 Antibody, KW-0761, in Cancer Patients. Clin Cancer Res 2015;21:4327-36. [Crossref] [PubMed]
- Berlato C, Khan MN, Schioppa T, et al. A CCR4 antagonist reverses the tumor-promoting microenvironment of renal cancer. J Clin Invest 2017;127:801-13. [Crossref] [PubMed]
- Tosolini M, Kirilovsky A, Mlecnik B, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res 2011;71:1263-71. [Crossref] [PubMed]
- Galon J, Angell HK, Bedognetti D, et al. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 2013;39:11-26. [Crossref] [PubMed]
- Marshall EA, Ng KW, Kung SH, et al. Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol Cancer 2016;15:67. [Crossref] [PubMed]
- Lee JH, Cho YS, Lee JY, et al. The chemokine receptor CCR4 is expressed and associated with a poor prognosis in patients with gastric cancer. Ann Surg 2009;249:933-41. [Crossref] [PubMed]
- Li JY, Ou ZL, Yu SJ, et al. The chemokine receptor CCR4 promotes tumor growth and lung metastasis in breast cancer. Breast Cancer Res Treat 2012;131:837-48. [Crossref] [PubMed]
- Liu Q, Rexiati M, Yang Y, et al. Expression of chemokine receptor 4 was associated with poor survival in renal cell carcinoma. Med Oncol 2014;31:882. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8. [Crossref] [PubMed]
- Deng L, Gyorffy B, Na F, et al. Association of PDCD1 and CTLA-4 Gene Expression with Clinicopathological Factors and Survival in Non-Small-Cell Lung Cancer: Results from a Large and Pooled Microarray Database. J Thorac Oncol 2015;10:1020-6. [Crossref] [PubMed]
- Forero A, Li Y, Chen D, et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol Res 2016;4:390-9. [Crossref] [PubMed]
- Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844-7. [Crossref] [PubMed]
- Braun M, Kirsten R, Rupp NJ, et al. Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system. Histol Histopathol 2013;28:605-10. [PubMed]
- Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653-64. [Crossref] [PubMed]
- Russell PA, Wainer Z, Wright GM, et al. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011;6:1496-504. [Crossref] [PubMed]
- Donnem T, Hald SM, Paulsen EE, et al. Stromal CD8+ T-cell Density-A Promising Supplement to TNM Staging in Non-Small Cell Lung Cancer. Clin Cancer Res 2015;21:2635-43. [Crossref] [PubMed]
- Schalper KA, Brown J, Carvajal-Hausdorf D, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 2015.107. [PubMed]
- Kinoshita T, Muramatsu R, Fujita T, et al. Prognostic value of tumor-infiltrating lymphocytes differs depending on histological type and smoking habit in completely resected non-small-cell lung cancer. Ann Oncol 2016;27:2117-23. [Crossref] [PubMed]
- Miyara M, Yoshioka Y, Kitoh A, et al. Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 2009;30:899-911. [Crossref] [PubMed]
- deLeeuw RJ, Kost SE, Kakal JA, et al. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res 2012;18:3022-9. [Crossref] [PubMed]
- Saito T, Nishikawa H, Wada H, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med 2016;22:679-84. [Crossref] [PubMed]


