Comparative study of MALDI-TOF MS and VITEK 2 in bacteria identification
Introduction
Traditionally, bacterial and fungal identifications in clinical microbiology laboratories are mainly carried out according to phenotype characteristics, including identifications of culture media, colony morphology, gram stain and various biochemical reactions (1). Although all of these methods can achieve high accuracies, it usually takes minimum one day or longer to complete the whole identification process. Molecular methods, such as real-time PCR, gene sequencing and microarray analysis, are quick methods for bacterial and fungal identification, but they come at a very high cost and require highly-trained technicians. Therefore, molecular methods are not routinely used for bacterial identification. A faster and easier technique for microbial identification would greatly enhance the conventional laboratory in providing more timely feedback to clinic. This is especially true in cases when patients are critically ill suffering from infectious diseases and where therapeutic intervention is urgently needed.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) can be used to obtain protein fingerprinting from whole bacterial cells (2). Through comparing these fingerprints to a reference database by the use of various algorithms, bacteria can be rapidly identified. The earliest application of this technique for bacterial identification dates back to 1975 (3), while the first related article was published in 1996 (Holland, et al.) (4). Studies in this field have been progressively advancing for the last decade (5). Until now, domestic comparative studies on mass spectrometry and other methods are relatively few. In order to compare the performance differences between MALDI-TOF MS and VITEK 2, the latter being the most-used automated identification technique in current microbiology laboratories, we employed the two systems in parallele to identify and analyze 1,025 isolates routinely isolated in 2012 at a microbiology laboratory of PLA General Hospital.
Methods
Bacteria isolates
A total of 1,025 isolates, composed of 1,020 bacterial strains and 5 fungal strains representing 25 different genera were selected for analysis. These bacteria were routinely isolated from clinical patients, such as Pseudomonas spp, Acinetobacter spp that cause lung infections, Enterobacteriaceae that cause blood and urinary tract infections. In addition, there are also some bacteria which grow slowly but have clinical significance such as Eikenella corrodens, Listeria monotytogens, Haemophilus infuenzae, etc.
VITEK 2 identification
GP, GN, YST, ANC and NH were selected to run identification analysis according to the different strains to be tested. The identification rate was expected to be above 93% by VITEK 2 (bioMérieux) (5), with quality control by stages via ATCC25922, ATCC27853 and ATCC25923.
MALDI-TOF MS identification
Strains were identified by MALDI-TOF MS using the VITEK MS system (bioMérieux). Loops were used to select and smear the subject isolates onto the sample spots on the target slide. Then 1 μL VITEK MS-CHCA matrix was applied over the sample and air dried until the matrix and sample co-crystallized. The target slide with all prepared samples then was loaded into the VITEK MS system to acquire the mass spectra of whole bacterial cell protein, mainly composed of ribosomal protein, for each sample. In the case of yeasts, 0.5 μL VITEK MS-FA was added to each sample on the target slide and allowed to air dry it before adding 1 μL VITEK MS-CHCA matrix. Finally, the mass spectra acquired for each sample were compared to the known mass spectra contained in the database for, and given a confidence score according to how close the acquired spectra matched those contained in the database. ATCC8739 was used as the quality control strain.
Gene sequencing
Discrepancies between VITEK 2 and MALDI-TOF MS were resolved by 16S rRNA gene sequencing. The results would be considered valid if the homologous rate was above 99%.
Standards for identification
In cases where the isolate was correctly identified at the species level or at the generic level by both VITEK MS and VITEK 2, the results were considered to be in accordance with one another. In instances that the two methods produced different results, or if the results from one method were not definite, 16S rRNA gene sequencing was used to resolve identification discrepancies. For isolates misidentified at the genus level, the results were considered a major mistake in the identification analysis. For isolates misidentified at species level but correctly identified at the genus level, the results were considered a minor mistake. Results not able to distinguish two different bacteria of the same genus also were considered as minor mistakes (6,7).
Results
Identification results by MALDI-TOF MS
Among the 1,025 total isolates, 1,021 (99.60%) isolates were accurately identified at the genus level and 957 (93.37%) isolates at the species level by MALDI-TOF MS.
Matched results by the two identification methods
Identification results for 949 (92.59%) isolates, belonging to 23 genera and 48 species, were in agreement (see Table 1) using both methods.
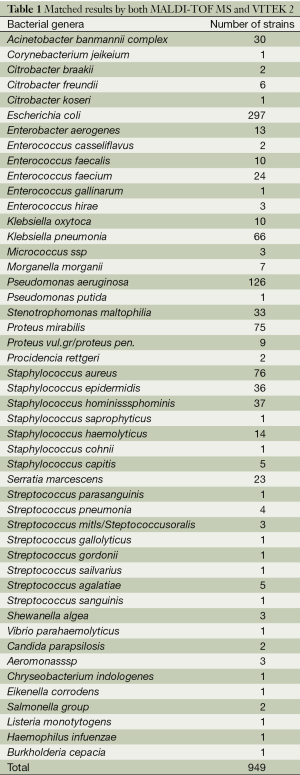
Full table
Unmatched results by the two identification methods
Among the 1,025 isolates, 76 (7.4%) isolates produced discordant results between the two identification methods. One strain had no definitive identification for each MALDI-TOF MS and VITEK 2. However, MALDI-TOF MS made no errors at the genus level while VITEK 2 made 6 (0.58%) errors at the genus level. At the species level, the identification error rates of the two methods were 5.56% and 6.24% for MALDI-TOF MS and VITEK 2 respectively (Table 2).
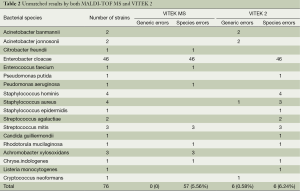
Full table
Discussion and Conclusions
In our laboratory, MALDI-TOF MS is an efficient, quick and relatively inexpensive per isolate method for identifying pathogenic microorganisms including bacteria and fungi. MALDI-TOF MS identification also is largely compatible with a large range of culture media and culture conditions, and is the fastest means to detect microbes in positive blood culture (8). Furthermore, MALDI-TOF MS has broader application prospects in the field of testing for drug resistance (9,10).
Shortening the turnaround time to pathogen identification and offering quicker results to the clinic remain the most important issues needed to be solved in the microbiology laboratory. Based on our data, experienced clinicians can treat critically infected patients appropriate antibiotic treatment given effective and timely identification of pathogenic bacteria. Shortening the turnaround time permits the clinician to treat patients faster, ultimately reducing the fatality rates, the length of patient stay in the clinic, and healthcare costs associated with patient care. MALDI-TOF MS offers one promising solution to shorten the turnaround time and relieve these pressures in the clinic.
In the present study, we chose 1,025 pathogenic bacteria routinely found in our laboratory for identification, and compared the performance of MALDI-TOF MS to the VITEK 2 system, the latter which is based on conventional biochemical identification that is presently found in most microbiology laboratories. Our results showed that MALDI-TOF MS offered higher identification accuracy and lower error rates at the species level when compared to VITEK 2. Additionally, MALDI-TOF MS dramatically shortened identification time from 6-8 hours to just a few minutes.
MALDI-TOF MS identified 99.60% of isolates to the genus level and 93.37% of isolates to the species level, which are slightly higher rates than those previously reported (5,7,11-14). Van Veen et al., for example, achieved accuracy rates of 97.1% and 92% to the genus and species levels, respectively. The difference in accuracy is most likely due to the different choices of strains. The strains in van Veen’s study included 61 yeast strains, among which 96.7% were identified to the genus level and 82.5% to the species level. Streptococcus viridans, streptococcus pneumoniae and anaerobic bacteria, all difficult pathogens to identify notably were included in van Veen’s study and not in our study. Most of the strains analyzed in our study were largely more commonly found pathogens, and the construction of the MALDI-TOF MS database may offer higher identification accuracies for these pathogens.
In conclusion, MALDI-TOF MS is a rapid, easy, relatively inexpensive and high-throughput method for identifying clinically relevant bacteria and fungi. MALDI-TOF MS offers very high accuracy, often higher than conventional methods, in identifying common microorganisms. Due to these key advantages and with continued development of MALDI-TOF MS technology and its clinical knowledgebase, MALDI-TOF MS will become a rapid, routine method for identifying pathogenic bacteria found in the conventional clinical microbiology laboratory.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Carroll KC, Weinstein MP. Manual and automated systems for detection and identification of microorganisms. In: Murray PR, Baron EJ, Jorgensen JH, et al. eds. Manual of clinical microbiology. 9th ed. Washington, DC: American Society for Microbiology Press, 2007:192-217.
- Fenselau C, Demirev PA. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom Rev 2001;20:157-71. [PubMed]
- Anhalt JP, Fenselau C. Identification of bacteria using massspectrometry. Anal Chem 1975;47:219-25.
- Holland RD, Wilkes JG, Rafii F, et al. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 1996;10:1227-32. [PubMed]
- Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009;49:543-51. [PubMed]
- Bizzini A, Durussel C, Bille J, et al. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol 2010;48:1549-54. [PubMed]
- van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol 2010;48:900-7. [PubMed]
- Drancourt M. Detection of microorganisms in blood specimens using matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a review. Clin Microbiol Infect 2010;16:1620-5. [PubMed]
- Burckhardt I, Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol 2011;49:3321-4. [PubMed]
- Hrabák J, Walková R, Studentová V, et al. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2011;49:3222-7. [PubMed]
- Blondiaux N, Gaillot O, Courcol RJ. MALDI-TOF mass spectrometry to identify clinical bacterial isolates: evaluation in a teaching hospital in Lille. Pathol Biol (Paris) 2010;58:55-7. [PubMed]
- Dupont C, Sivadon-Tardy V, Bille E, et al. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect 2010;16:998-1004. [PubMed]
- Dubois D, Leyssene D, Chacornac JP, et al. Identification of a variety of Staphylococcus species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2010;48:941-5. [PubMed]
- Spanu T, De Carolis E, Fiori B, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to rpoB gene sequencing for species identification of bloodstream infection staphylococcal isolates. Clin Microbiol Infect 2011;17:44-9. [PubMed]


