Sputum purulence-associated microbial community compositions in adults with bronchiectasis
Introduction
Bronchiectasis is a debilitating chronic inflammatory airway disease which arises from multiple etiologies. The interplay among the etiology, altered microenvironment and defective host-defense may have predisposed to recurrent chest infections of pathogens (1,2) that correlate with bronchiectasis exacerbations and increased mortality. Sputum production, which results from mucus hypersecretion and exaggerated airway inflammation, is one of the major complaints among symptomatic bronchiectasis patients (3) whose sputum bacterial compositions are heterogeneous (4,5). Clinically, sputum purulence has been an important parameter reflecting the magnitude of airway inflammation and infection. Sputum purulence, which is commonly rated by using color charts in clinical settings, has been correlated with sputum myeloperoxidase and neutrophil elastase activity and pathogen (i.e., pathogenic bacteria) infections (6,7).
Conventionally, physicians prescribed antibiotics based on sputum purulence in conjunction with assessment of clinical symptoms such as sputum volume and cough frequency. Nonetheless, antibiotics are frequently prescribed to bronchiectasis patients (particularly those with purulent sputum) regardless of culture findings in many developing countries (such as China). Some patients with purulent sputum tested negative for bacterial culture, suggesting that culture techniques have limited value for identifying pathogenic bacteria. Little is known regarding bacterial compositions associated with purulent sputum in bronchiectasis. Studies that comprehensively characterize microbial compositions according to purulence are needed to inform physicians in which subgroup should antibiotic prescription be prioritized.
We hypothesized that sputum purulence partially correlated with the sputum microbial compositions and that sputum purulence was not solely responsible for interpreting the variations in microbial compositions in patients with clinically stable bronchiectasis and during bronchiectasis exacerbations. In this study, we sought to compare bacterial compositions between purulent and non-purulent sputum, and between culture-positive and -negative purulent sputum, followed by comparison of changes in bacterial diversity during exacerbations with 16srRNA sequencing (8,9).
Methods
Study participants
Adults with bronchiectasis confirmed with high-resolution computed tomography (effective within 12 months, reviewed by an experienced radiologist) were consecutively recruited between March 2014 and November 2015. The criteria for diagnosing bronchiectasis have been published previously (3). Patients were free from exacerbations for >4 weeks (4). We excluded patients with malignancy, upper airway bacterial/viral infection, or antibiotic use (except for the low-dose maintenance therapy with macrolides) within 4 weeks. Our local ethics committee gave approval Medical Ethics Year 2012 (The 33rd), and patients signed informed consent.
Study design
Patients underwent baseline visits when stable. For those with exacerbations who contacted investigators, exacerbation visits were scheduled, followed by 14-day antibiotic therapy as recommended by the international guidelines with minor changes (3). In this study, bronchiectasis exacerbations denoted continuous (48 h or greater) significant worsening of 3 or more symptoms or signs: significantly increased cough frequency, sputum volume, sputum purulence, dyspnea, fever, fatigue/malaise, and new-onset or worsening of haemoptysis (3,4). Convalescence visits were scheduled 1 week after antibiotic therapy. At each visit, assessment included history inquiry, spirometry and sputum sampling. Spontaneous sputum plugs (the most purulent portion of the sample) which met quality control (white blood cell: epithelial cell ratio >2.5:1) were split for culture (blood/chocolate agar for overnight incubation, targeting at Pseudomonas aeruginosa, Haemophilus spp., and other pathogenic bacteria) (4) and sequencing. Purulence was evaluated by an investigator masked to patient’s profiles, with purulent sputum denoting purulence scores (scale: 0–8) of 6 or greater (0: absence of sputum; 1: completely transparent; 2: almost transparent; 3: translucent but colorless; 4: opaque and milky white; 5: grey; 6: pale green; 7: moderately green; 8: dark green sputum) (10).
16srRNA sequencing
Nucleic acids were extracted using physical disruption (vortex) and centrifugal absorption column (HiPure DNA Kit B, Magen Inc., Guangzhou, China). Quality control of the samples was achieved via agarose gel electrophoresis and ND-100 Nanodrop system (Thermo Fisher Inc., USA). Qualified samples were subject to library construction with TruSeqTM Custom Amplicon Sample Prep Kit (Illumina Inc., USA), and barcoded and mixed before pooling. The quality of the library was assessed with Agilent 2200 TapeStation (Agilent Inc., USA) and Qubit 2.0 (ThermoFisher Inc., USA). 2.1nM Phix Control (19:1) and sodium hydroxide were added for incubation of the sample, followed by dilution with HT1 solution (1:100). We performed DNA sequencing with Miseq System (Illumina Inc., USA). Raw reads were denoised, followed by chimera removal. High-quality sequences were clustered into operational taxonomic units (OTUs) at 97% similarity with UCLUST software. The sequences for individual OTU were aligned with BLASTn, and the taxonomic identities were assigned by using Ribosomal Database Project classifier (version 2.2). The QIMME software was employed for rarefaction of the denoised files, which enabled subsequent calculation of the relative abundance and the diversity metrics. Sequences were deposited in GenBank under accession number SAMN06768146-SAMN06768292.
Statistical analysis
We performed analyses with R package (www.r-project.org) and Graphpad Prism 5.0 (Graphpad Inc., San Diego, USA). We classified patients into the group with mucoid or mucopurulent sputum (M+MP, n=16), culture-negative (no pathogenic bacteria) purulent sputum (C−P+, n=16), and culture-positive purulent sputum (C+P+, n=73). Group M+MP was not stratified because of limited sample sizes. One-way analysis-of-variance or Kruskal-Wallis test was applied for the comparison of three groups for continuous variables, followed by least-square difference test for two-group comparisons. We used independent t-tests or Wilcoxon signed-rank tests to compare the continuous variables between the two groups. Paired t-tests were conducted for the pairwise sputum samples. Categorical variables were compared with chi-square test. We compared the relative abundance and Shannon-Wiener diversity index (an alpha-metric reflecting the number of types of sequences within any individual sample) to reflect the magnitude of dysbiosis (altered microbial compositions in response to external stimuli such as disease). We ran similarity of percentage analysis with Community Analysis Package version 5.3.3.472 (Hampshire, UK).
Results
Of 207 patients screened, 105 with spontaneous sputa had available purulence scores (64 patients declined and 38 produced insufficient sputum). Median purulence score was 6.0 (range, 1.0–8.0). 85 patients cultured positive to pathogenic bacteria (42.9% isolated Pseudomonas aeruginosa). Idiopathic (32.3%) and post-infectious (37.1%) were the most common etiologies. Twenty five percent, 25.0% and 12.3% of the patients in M+MP, C−P+, and C+P+ group had been prescribed with inhaled corticosteroids within 6 months (P>0.05), respectively. No patients were receiving oral/inhaled antibiotics. Bronchiectasis Severity Index was significantly higher in group C+P+ (mean: 6.8) than in groups M+MP and C−P+ (mean: 3.8 and 4.8, P=0.005) (Table 1). Twenty-two patients attended exacerbation visits (5, 4 and 13 in group M+MP, C−P+, and C+P+, respectively), of whom 20 participated in convalescence visits.
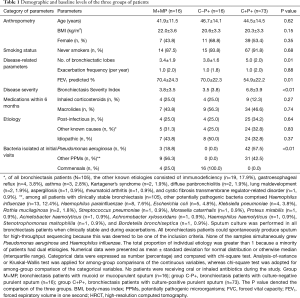
Full table
We detected a mean of 42, 43 and 36 unique OTUs in groups M+MP, C−P+ and C+P+, respectively. Overall, Proteobacteria was the dominant phylum among all patients, with greater sputum purulence score correlating with higher relative abundance (Figure 1A). At genera levels, Pseudomonas dominated among all patients, with greater sputum purulence score correlating with higher relative abundance (Figure 1B). Group M+MP demonstrated significantly lower abundance of Proteobacteria (78.6% vs. 84.6%) and higher abundance of Firmicutes (10.9% vs. 6.7%) than the rest combined (both P<0.05). Group C−P+ had significantly lower abundance of Proteobacteria (64.2% vs. 89.1%) and higher abundance of Firmicutes (11.5% vs. 5.7%) than group C+P+ (both P<0.01). Group C−P+ had significantly higher Shannon-Wiener diversity index (0.902 vs. 0.393) compared with group C+P+ (P<0.05) (Figure 2A,B). Proteobacteria consistently contributed most to community similarity in groups M+MP, C−P+ and C+P+ (85.8%, 74.6% and 94.9%). However, Firmicutes contributed considerably to community similarity in groups M+MP (8.4%) and C−P+ (11.7%).
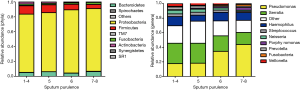
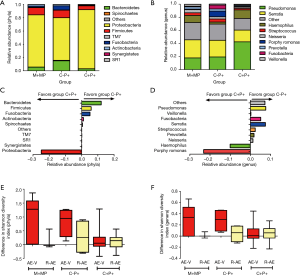
Pseudomonas spp. predominated in group C+P+, whereas Serratia spp. predominated in groups M+MP and C−P+. Group M+MP demonstrated significantly lower abundance of Pseudomonas (17.8% vs. 38.3%) and higher abundance of Serratia spp. (27.8% vs. 19.3%) than the rest combined (both P<0.05). Group C-P+ yielded markedly lower abundance of Pseudomonas spp. (19.3% vs. 42.5%) than group C+P+ (P<0.01). However, the abundance of Serratia spp. was comparable (25.4% vs. 18.0%). Group C−P+ demonstrated significantly higher Shannon-Wiener diversity index (2.120 vs. 1.381) than group C+P+ (P<0.05) (Figure 2C,D). Pseudomonas spp. contributed most to community similarity in group C+P+ (49.7%), but not in groups M+MP (19.2%) and C−P+ (19.9%). Serratia spp. (21.6–31.8%) contributed considerably to community similarity in bronchiectasis.
Although groups C−P+ plus C+P+ differed considerably from group M+MP in microbial compositions, sputum purulence scores correlated with the abundance of Proteobacteria and most genera (including Pseudomonas spp.) and Shannon-Wiener diversity index, at phyla and genera levels (data not shown).
We compared microbial compositions of sputum samples when clinically stable and during exacerbation, and from exacerbation to convalescence. A trend towards greater differences in Shannon-Wiener diversity index was observed in groups M+MP and C−P+ at phyla and genera levels (Figure 2E,F). No significant differences were found in the abundance of Proteobacteria and Pseudomonas spp. when comparing paired samples collected when clinically stable and during exacerbations (P>0.05).
Discussion
In this study, we have demonstrated that bronchiectasis patients with purulent sputum and isolation with pathogenic bacteria had considerably greater magnitude of airway dysbiosis, particularly when assessed in terms of the relative abundance of Proteobacteria and the alpha-diversity index. We have confirmed some of the findings from a previous study that sputum purulence was a strong predictor of potentially pathogenic microorganisms (PPM) infection (11). Furthermore, our study partially echoed the findings that sputum purulence correlated with pathogenic bacterial infection in hospitalized patients with COPD exacerbations (12). Nonetheless, our study has further revealed a considerable heterogeneity of microbial compositions even within the subgroups classified according to sputum culture findings and purulence. Notably, sputum microbial compositions do not solely depend on sputum culture positivity. A considerable heterogeneity in microbial compositions could be found among individual bronchiectasis patients within the same subgroup (i.e., culture-positive or culture-negative group) (data not shown).
Focusing solely on purulent sputum in clinically stable bronchiectasis, microbial compositions varied substantially among individuals (e.g., the abundance of Proteobacteria and Pseudomonas spp. was 8.6~84.6% and 1.5~92.2%). Only those with purulent culture-positive sputum, which demonstrated minimal changes in bacterial diversity during exacerbations, displayed the most prominent degree of airway dysbiosis. These findings reaffirmed that sputum purulence alone was not the sole dependent factor that influences on microbial compositions. Importantly, our study suggested that stratification of patients based on the combination of sputum purulence and culture findings may help better elucidate the characteristics of airway dysbiosis and explore the possible source of variation in sputum microbial compositions. Therefore, sputum purulence and culture findings might have complementary value for guidance of antibiotics prescription in the clinical settings. In some patients, the minor degree of airway dysbiosis indicated the alternative sources of sputum purulence (13), such as exaggerated elastase release. According to another study, outgrowth of Proteobacteria and/or Pseudomonas spp. contributes to more prominent dysbiosis (14), whereas Haemophilus spp. and Serratia spp. counter the expansion of Pseudomonas spp. Our study has partially corroborated with these findings, and has further indicated the potential association between sputum purulence and outgrowth of Proteobacteria and/or Pseudomonas spp. in some, but not all, of the purulent sputum samples.
Interestingly, the changes in the relative abundance of Proteobacteria were greater in group M+MP compared with groups C−P+ and C+P+ during bronchiectasis exacerbations as compared with clinically stable visits. It is likely that the minor magnitude of airway dysbiosis (lower relative abundance of Proteobacteria that resulted in minor suppression of other PPMs) may be responsible for the greater changes in microbial composition during exacerbations. The minor changes in microbial compositions during exacerbations as compared with clinically stable visits have been reported in a previous study (8). However, because the sputum purulence assessment was not performed, we were unable to directly compare the findings further. According to the study by Purcell et al. (5), there existed a considerable difference in the patterns of changes in sputum microbial compositions during bronchiectasis exacerbations compared with clinically stable visits. Notably, the presence of Burkholderiales, Pasteurellaceae, Streptococcaceae and some other PPMs correlated with bronchiectasis exacerbations. In our study, we did not specifically focus on the comparison of changes in all bacterial phyla and genera, therefore we were unable to address whether the presence of the bacteria mentioned above might help interpret the greater change in microbial compositions during exacerbations in the group M+MP. More mechanistic investigations of the factors that drive the major changes in sputum microbial compositions during exacerbations are needed.
There are some limitations that should be considered. Some of the subgroup analysis might be not sufficiently powered because of the limited sample sizes. The sample size for the subgroups was unbalanced. Patients aged on average lower compared with the populations in other geographic regions. Our findings might not be completely extrapolated to patients in other countries because of the high relative abundance of Proteobacteria and Serratia and the low relative abundance of Haemophilus spp. We did not measure clinically relevant markers (i.e., neutrophil elastase activity) that have been correlated with sputum purulence. Finally, bronchiectasis exacerbations are highly heterogeneous events which indicated significant variations in sputum microbial compositions that cannot be adequately addressed based on the current study design.
This study documented the significant heterogeneity of purulent sputum in bronchiectasis. Culture findings coupled with sputum purulence assessment may better guide clinicians for antibiotics prescription.
Acknowledgements
We thank Bei-Qing Kuang, Xiu-Juan Tang, Mei Zheng, Zi-Qing Ye, Ming-Feng Li, Qian Li, Zhi-Wen Chen, Zhi-Qiang Huang, Fei-Long He, Xiao-Yong Shen, Chao Wen, and Prof. Bi-liang Zhang (Guangzhou Ribobio Co. Ltd., Guangzhou, China) for their technical assistance and advice.
Footnote
Conflicts of Interest: Dr. Guan declared that he has received National Natural Science Foundation No. 81870003, Pearl River S&T Nova Program of Guangzhou No. 201710010097, and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme 2017. Dr. Gao declared that he has received National Natural Science Foundation No. 81500006. Profs. Zhong and Chen declared that they had received Changjiang Scholars and Innovative Research Team in University ITR0961, The National Key Technology R&D Program of the 12th National Five-year Development Plan 2012BAI05B01 and National Key Scientific & Technology Support Program: Collaborative innovation of Clinical Research for chronic obstructive pulmonary disease and lung cancer No. 2013BAI09B09. Other authors have no conflicts of interest to declare.
Ethical Statement: Our local ethics committee gave approval Medical Ethics Year 2012 (The 33rd), and patients signed informed consent.
References
- Polverino E, Goeminne PC, McDonnell MJ, et al. European Respiratory Society guidelines for the management of adult bronchiectasis. Eur Respir J 2017.50. [PubMed]
- Chen ZG, Li YY, Wang ZN, et al. Aberrant epithelial remodeling with impairment of cilia architecture in non-cystic fibrosis bronchiectasis. J Thorac Dis 2018;10:1753-64. [Crossref] [PubMed]
- Pasteur MC, Bilton D, Hill AT, et al. British Thoracic Society guideline for non-CF bronchiectasis. Thorax 2010;65:i1-58. [Crossref] [PubMed]
- Guan WJ, Gao YH, Xu G, et al. Sputum bacteriology in steady-state bronchiectasis in Guangzhou, China. Int J Tuberc Lung Dis 2015;19:610-9. [Crossref] [PubMed]
- Purcell P, Jary H, Perry A, et al. Polymicrobial airway bacterial communities in adult bronchiectasis patients. BMC Microbiol 2014;14:130. [Crossref] [PubMed]
- Goeminne PC, Vandooren J, Moelants EA, et al. The Sputum Colour Chart as a predictor of lung inflammation, proteolysis and damage in non-cystic fibrosis bronchiectasis: a case-control analysis. Respirology 2014;19:203-10. [Crossref] [PubMed]
- Stockley RA, Hill SL, Morrison HM, et al. Elastolytic activity of sputum and its relation to purulence and to lung function in patients with bronchiectasis. Thorax 1984;39:408-13. [Crossref] [PubMed]
- Tunney MM, Einarsson GG, Wei L, et al. The lung microbiota and bacterial abundance in patients with bronchiectasis when clinically stable and during exacerbation. Am J Respir Crit Care Med 2013;187:1118-26. [Crossref] [PubMed]
- Stockley RA, Hill SL, Morrison HM, et al. Elastolytic activity of sputum and its relation to purulence and to lung function in patients with bronchiectasis. Thorax 1984;39:408-13. [Crossref] [PubMed]
- Tsang KW, Tan KC, Ho PL, et al. Inhaled fluticasone in bronchiectasis: a 12 month study. Thorax 2005;60:239-43. [Crossref] [PubMed]
- Miravitlles M, Kruesmann F, Haverstock D, et al. Sputum colour and bacteria in chronic bronchitis exacerbations: a pooled analysis. Eur Respir J 2012;39:1354-60. [Crossref] [PubMed]
- Soler N, Esperatti M, Ewig S, et al. Sputum purulence-guided antibiotic use in hospitalised patients with exacerbations of COPD. Eur Respir J 2012;40:1344-53. [Crossref] [PubMed]
- Goeminne PC, Vandooren J, Moelants EA, et al. The Sputum Colour Chart as a predictor of lung inflammation, proteolysis and damage in non-cystic fibrosis bronchiectasis: a case-control analysis. Respirology 2014;19:203-10. [Crossref] [PubMed]
- Rogers GB, van der Gast CJ, Serisier DJ. Predominant pathogen competition and core microbiota divergence in chronic airway infection. ISME J 2015;9:217-25. [Crossref] [PubMed]


