
Expanding the gamut of circulating tumor DNA applications
Historically, advanced non-small cell lung cancer (NSCLC) has been a devastating cancer diagnosis with an aggressive course and poor prognosis despite extensive and toxic therapy, however with the advent of personalized therapy targeting molecular aberrancies, the field of lung cancer has made significant advances in outcomes with concomitant improvements in toxicity profiles (1,2). For those patients who are candidates for molecularly targeted therapy, identification of mutations by tumor tissue testing has become the gold standard (3,4). A number of limitations to tissue specimen analysis exist, however, that make it less practical in certain cases including inadequate quantity of specimen and poor-quality specimen. What makes tumor testing even more challenging is its invasiveness and the inability to monitor dynamic changes throughout treatment which has become a key issue with the recognition of molecular mechanisms of acquired resistance permitting new generation treatment options. Recently, circulating tumor DNA (ctDNA) testing in plasma has emerged as an alternative means of identifying and monitoring epidermal growth factor receptor (EGFR) mutations, and it is currently approved by the Food and Drug Administration (FDA) for EGFR mutation detection when tumor tissue collection is inadequate (5,6). Plasma based testing of EGFR mutations can alleviate many of the issues that arise with tissue-based testing, however further studies to clarify the relationship between liquid and tissue biopsy and its effect on therapeutic efficacy are needed. In multiple retrospective studies a correlation has already been established between blood-based EGFR mutations and EGFR-tyrosine kinase inhibitor (TKI) efficacy suggestive of excellent positive and lesser negative predictive value, yet a prospective trial to adequately determine treatment outcomes based upon plasma-based treatment assignment has been lacking until recently (7-12).
It is in this context that the recent BENEFIT trial has attempted to fill this clinically important void. This study is a prospective, single arm phase 2 clinical trial conducted by Wang and colleagues in which they investigate the use of ctDNA-based EGFR mutation testing to guide diagnostic and therapeutic decisions in patients with advanced lung adenocarcinoma receiving first line EGFR TKIs. The study recruited treatment naïve patients with stage IV metastatic lung adenocarcinoma whose ctDNA testing measured by droplet digital polymerase chain reaction (PCR) (ddPCR) showed positive results for an actionable EGFR mutation, and they were administered first line oral gefitinib treatment. Primary endpoint was objective response while secondary endpoints included median progression-free survival (PFS) and safety. Next-generation sequencing (NGS) was also performed on all blood samples to assess for additional mutational analysis and EGFR ddPCR testing was repeated at 8 weeks to allow exploratory studies on correlation of outcome with ctDNA clearance. EGFR testing was also performed on available tissue specimens to allow correlations between the two platforms.
The authors as anticipated found a high specificity (93.9%) and positive predictive value (95.8) between plasma and tissue derived EGFR mutational status, and a sensitivity of 70%. Of note is that this low sensitivity is not a surprise; depending on tumor type, volume and proclivity for ctDNA shedding, different rates of detectability can be expected with 70% being a fairly typical figure in the setting of advanced NSCLC. Among 188 patients who were enrolled, the objective response rate was 72.1% and median PFS was 9.5 months which is comparable to results from tissue detection-based clinical trials with the use of gefitinib providing clinical validation for ctDNA based subject selection. Indeed, as ctDNA tends to be positive in higher tumor burden patients, the average outcome of a patient population selected by ctDNA testing might be expected to be inferior as compared to tissue based detection where the subset includes low tumor burden, non-shedder tumors as well. Additionally, subgroup analysis provided intriguing information about which groups of patients may do better with EGFR TKI therapy. Among the patients with de novo Thr790Met mutations, objective response was lower (33% vs. 74.1%) and median PFS was shorter (5.6 vs. 9.6 months), a result expected given the use of gefitinib rather than osimertinib. More interestingly, a higher percentage of patients with EGFR exon 19 deletions had objective response compared with exon 21 Leu858Arg although there was no significant difference between median PFS.
Next, the authors divided the patients based on NGS results into the following three subgroups: (I) EGFR-sensitizing mutations alone vs. (II) EGFR-sensitizing mutations plus tumor-suppressor gene mutations vs. (III) EGFR-sensitizing mutations plus oncogenic driver mutations. Median PFS was longest in the first group and shortest in the third group (13.2, 9.3, 4.7 months, respectively). The key issue here is that baseline NGS is presumed to reflect higher genomic heterogeneity thereby leading to either lesser dependence on the EGFR oncogenic pathway leading to primary resistance or earlier development of acquired resistance. These findings are in line with recent reports by the Lung Cancer Mutation Consortium showing significant impact of baseline p53 alterations on outcome (13).
The most intriguing findings, however, came from the analysis when Wang et al. assessed dynamic changes in EGFR mutations throughout therapy. They found that in patients whose plasma EGFR ctDNA cleared by 8 weeks, median PFS was prolonged significantly. In addition, it appeared that ctDNA cleared at a higher rate by week 8 in patients with exon 19 deletions as compared to exon 21 Leu858Arg mutations, and in those with exon 19 deletions whose deletion cleared at week 8, these patients were noted to have lesser abundance of mutant DNA at baseline in comparison with those with exon 19 deletions who did not achieve clearance. Overall, subjects with ctDNA clearance had significantly less circulating DNA tumor burden in essence.
These findings are quite provocative and have potentially broad diagnostic and therapeutic implications, although not without raising some concern about the appropriate use of cell free DNA testing. Of the 260 patients who had tissue biopsy positive for EGFR mutations, 78 patients (30%) had ctDNA test negative for the corresponding mutation. Moreover, 8 patients had EGFR mutations detected in ctDNA, but not in tumor tissue, and this group of patients had lower median PFS (6 months). Although the rigorous cutoff described by the authors was offered as an explanation for the lower sensitivity between ctDNA detected and tumor tissue detected EGFR mutations, these inconsistencies along with the lower PFS in patients with only blood-based EGFR mutation detection indicate that clinicians should be cautious in interpreting the data of ctDNA, and thereby complete reliance on upfront ctDNA testing for EGFR or other molecular analysis remains inappropriate given its limited negative predictive value. Plasma ctDNA testing should be a complement and not a replacement to tissue-based analysis when available.
Despite the limitations listed, Wang and colleagues should be commended for conducting an impressive and elegant study whose results significantly add to the available body of literature while raising several relevant questions that may foster further avenues of research. The most appealing potential application of ctDNA is its use as a predictive biomarker, identified in this trial by both the correlation of prognosis with level of ctDNA abundance at diagnosis and of treatment response correlating with dynamic changes in levels throughout therapy. Future research to better flesh out this role would be of high yield and could make significant impact in other areas as well; for example, monitoring of patients after completion of definitive therapy and assessing benefits of immunotherapy where imaging at times might be less reliable. Other directions for further investigation include the use of ctDNA testing in patients with uncommon EGFR mutations, which was not assessed in this trial, the therapeutic implications from data that suggests patients with exon 21 Leu858Arg, tumor suppressor or oncogenic mutations fare worse with EGFR TKI therapy, and clarification of guidelines for NGS panel testing for the upfront management of advanced NSCLC, not only in regard to EGFR testing, but also for assessment of multiple other markers including ALK, ROS, and B-Raf among others.
Besides ddPCR, there are now various commercial assays available including real-time PCR [Cobas and Amplification Refractory Mutation System (ARMS)], other digital platforms [Beads, Emulsions, Amplification and Magnetic (BEAMing)], NGS, and most recently, a possibly even more sensitive type of ddPCR that has been developed called a library aliquot-based droplet digital PCR (LAB-ddPCR) (14-16). While these assays have variable sensitivities and specificities with further standardization to be called for to allow cross platform comparisons, Wang et al. add significant data to further underscore the clinical utility and validity of ctDNA testing in the management of patients with advanced NSCLC.
It is still ill-defined which patients should undergo panel testing and if this can be done by plasma-based NGS, which within this study was not confirmed with tissue NGS leaving the validity of their results in question. Of note, there are ongoing studies currently recruiting patients to further investigate the association between liquid and tissue biopsy in lung NSCLC, the clonal status of sensitizing mutations with response to targeted therapy, and the effect that sensitizing and resistance mutations and its abundance has on prognosis and further clarity on this subject is anticipated. A recently published study in the Annals of Oncology attempted to answer the question of plasma-based NGS validity by conducting a blinded analysis of amplicon-based plasma NGS in advanced NSCLC compared with plasma ddPCR and tumor genotype (17). They found that amplicon-based plasma NGS was able to detect a full range of targetable genotypes including fusion genes with high accuracy, suggesting that plasma NGS could be an acceptable method of genomic analysis, although which assay is superior remains unclear. Lastly, it should be stated that while the BENEFIT study used gefitinib, a first-generation EGFR TKI, the recently published pivotal FLAURA trial has now similarly shown that outcomes with the third generation TKI, osimertinib in the front-line setting in patients with EGFR-mutated advanced NSCLC are similar in subjects with ctDNA positive disease as compared to the overall patient population again underscoring the clinical validity of ctDNA-based patient selection for EGFR TKI therapy (18).
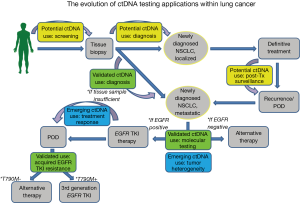
In summary, ctDNA analysis is a very exciting and welcome new addition to the armamentarium of clinically relevant and validated lung cancer biomarkers with tremendous potential to change therapeutic guidelines for patients, better tailor and individualize therapy, improve outcomes, and provide quicker, less invasive methods for diagnosis thus allowing for greater access to patients (Figure 1). However, while ctDNA testing is fully validated in the context of upfront EGFR mutation detection and EGFR T790M testing, more research needs to be done to clarify the role of ctDNA testing in other settings. It is our hope that studies like this will galvanize even more research on this topic and soon we will have a more comprehensive understanding of how to incorporate plasma ctDNA into practice throughout the treatment continuum.
Acknowledgements
None.
Footnote
Conflict of Interests: BH has received research funding from Merck, Mirati, Novartis, Pfizer, Takeda, Eli-Lilly, Boehringer-Ingelheim, BMS, Astra-Zeneca, AbbVie and has received consulting fees from Guardant Health, Foundation Medicine, Astra-Zeneca, Genentech, Boehringer-Ingelheim, Novartis, Pfizer, Spectrum, Ignyta, Eli-Lilly, Merck, BMS. The other author has no conflicts of interest to declare.
References
- Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- Lee CK, Brown C, Gralla RJ, et al. Impact of EGFR inhibitor in non-small cell lung cancer on progression-free and overall survival: a meta-analysis. J Natl Cancer Inst 2013;105:595-605. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med 2013;137:828-60. [Crossref] [PubMed]
- Leighl NB, Rekhtman N, Biermann WA, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J Clin Oncol 2014;32:3673-9. [Crossref] [PubMed]
- Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650-61. [Crossref] [PubMed]
- Singh AP, Li S, Cheng H. Circulating DNA in EGFR-mutated lung cancer. Ann Transl Med 2017;5:379. [Crossref] [PubMed]
- Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009;27:2653-9. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Han B, Tjulandin S, Hagiwara K, et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 2017;113:37-44. [Crossref] [PubMed]
- Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Reck M, Hagiwara K, Han B, et al. ctDNA Determination of EGFR Mutation Status in European and Japanese Patients with Advanced NSCLC: The ASSESS Study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Aisner DL, Sholl LM, Berry LD, et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 2018;24:1038-47. [Crossref] [PubMed]
- Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer Med 2017;6:154-62. [Crossref] [PubMed]
- Zhang R, Chen B, Tong X, et al. Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA. Cancer Manag Res 2018;10:1209-18. [Crossref] [PubMed]
- Yang K, Li J, Zhao J, et al. Developing Ultrasensitive Library-Aliquot-Based Droplet Digital PCR for Detecting T790M in Plasma-Circulating Tumor DNA of Non-small-Cell-Lung-Cancer Patients. Anal Chem 2018;90:11203-9. [Crossref] [PubMed]
- Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol 2018;29:1049-55. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]


