
Advance of theragnosis biomarkers in lung cancer: from clinical to molecular pathology and biology
Lung cancer is the leading cause of cancer deaths in men and women worldwide (1). Historically, it was divided in two major groups—small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) with 80% to 85% of cases belonging to the latter group (2). Over the last years, however, the advent of next-generation sequencing (NGS) technologies and sequencing studies revealed that this classification is outdated and that particularly the term “non-small cell lung cancer” involves an extremely heterogeneous set of diseases at the molecular level that translates into both, tumors’ clinical behavior and therapeutic decision-making (3).
A specific molecular subtype of NSCLC is defined by rearrangements of the anaplastic lymphoma kinase (ALK), which is found in about 4% of patients with lung adenocarcinomas (4) predominantly at younger age and mostly in light- or never-smokers (5). Histologically, it is often, but not exclusively, characterized by poor-differentiation with acinar-predominant structure and a mucin/signet-ring cell pattern (6,7).
The ALK gene encodes a receptor tyrosine kinase, which plays a significant role in the development and function of the nervous system, where it controls the basic mechanisms of cell proliferation, survival, and differentiation in response to extracellular stimuli (8). Its activation results in dimerization and autophosphorylation of the kinase domain consequently activating downstream signaling pathways, such as the RAS/MAPK, PI3K/AKT, and JAK/STAT pathways (9). ALK is considered an orphan receptor with unknown ligands, however, several factors have been described to interact with the receptor and activate ALK downstream signaling (10-13). Under physiological conditions, ALK is only embryonically expressed in early developmental regulation but not in the adult lung.
Under pathologic conditions, such as ALK translocated-NSCLC, the ALK gene brakes and fuses with diverse partners, leading to a constitutively activated kinase resulting in the uncontrolled activation of its downstream signaling pathways. These signaling cascades can contribute to uncontrolled cell proliferation, survival migration, angiogenesis and metastasis (14).
ALK fusions arise from breakage of the ALK gene, located on chromosome 2, and subsequent fusion of the 3' end of ALK, and the 5' portion of a different gene providing its promoter. The ALK breakpoint lies most frequently within intron 19 and, rarely, within exon 20, preserving the tyrosine kinase domain. The most common fusion partner is EML4 (echinoderm microtubule-associated protein-like 4), however, multiple different 5' translocation partners have been identified, including KIF5B, KLC1, TFG, TPR, HIP1, STRN, DCTN1, SQSTM1, BIRC6 (15). EML4 and ALK each map to the short arm of chromosome 2 but have opposite orientations, due to inversion of EML4 (16). The precise underlying mechanisms triggering the development of ALK gene rearrangements are not fully understood yet. In contrast to ALK, the EML4 breakpoints differ frequently, defining different variants of the EML4-ALK fusion gene. To date, more than 15 EML4-ALK variants have been identified with some variants being expressed as multiple isoforms, as reviewed in (17). All variants share the intracellular kinase domain of ALK, and contain the trimerization domain of EML4, which is required for the constitutive activation of ALK through oligomerization and autophosphorylation (18).
For the detection of ALK rearrangements in the routine clinical setting most laboratories use IHC (immunohistochemistry) and/or FISH (fluorescence in situ hybridization), which is cost effective and easily applicable with the disadvantage that other fusion partners than EML4 cannot be determined. However, molecular approaches, such as RNA-sequencing analyses by Archer®FusionPlex® have recently emerged as relevant alternatives and may provide an effective and accurate alternative to FISH testing for the detection of both, known and novel ALK rearrangements in clinical diagnostic settings (19).
In 2010, Kwak et al. were the first to show that the inhibition of ALK in ALK+-lung tumors by the multi-targeted tyrosine kinase inhibitor (TKI) crizotinib resulted in tumor shrinkage or stable disease in most patients (20), and only one year later, crizotinib was approved by the FDA as the first licensed ALK inhibitor for ALK+-NSCLC (21). Despite the enormous efficacy of crizotinib, resistance continued to be a major impediment.
Due to recent analyses of post-treatment tumor tissue samples by Camidge et al. (22) our knowledge in terms of molecular resistance mechanisms to ALK inhibitors has been significantly extended (22). The authors defined two types of ALK-inhibitor resistance: the ALK-dominant type (also known as “on-target” type) represents 50% of cases and is defined by secondary mutations within the ALK kinase domain or gene amplifications of the ALK gene. The non-dominant type is characterized by the activation of ALK-bypassing oncogenic pathways, including EGFR and KRAS mutations (23), amplification of KIT (24) or activation of the IGF1R pathway (25). Further described resistance mechanisms are epithelial-mesenchymal transition (EMT) (26) and autophagy (27).
With increasing knowledge over the last years, therapeutic options for advanced ALK+-NSCLC have been continuously improved and increasingly potent and selective ALK inhibitors (e.g., ceritinib and alectinib) have been approved by the FDA (14). A recent phase III trial revealed that the second-generation ALK inhibitor alectinib showed superior efficacy and lower toxicity in primary treatment of ALK-positive NSCLC and was further associated with activity against CNS (28). Consequently, it outperformed the first approved ALK inhibitor crizotinib and has recently been used as first line therapy. Further next-generation inhibitors, such as brigatinib and lorlatinib are currently under development (29,30). Of note, in the past few years, the “traditional” strategy of sequential treatment approaches, in which ALK+-patients initially received first-generation TKIs, which were replaced by next-generation TKIs and/or chemotherapy upon disease progression, has been challenged and tended to shift to the use of next-generation TKIs in the frontline setting (31).
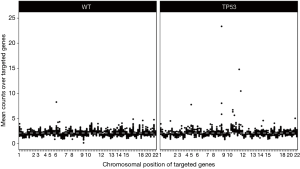
Despite the well-advanced elucidation of resistance mechanisms, it remains unclear why some patients relapse faster or show worse up-front response to ALK inhibition treatment. Recent data by Lin et al. showed that specific EML4-ALK variants may be associated with the development of resistance mutations to ALK TKIs in ALK+-NSCLC (32). In 2016, Gainor et al. performed the largest series of repeat biopsies (with 103 cases being studied) from patients with TKI-resistant ALK+-NSCLC, using a combination of genetic sequencing, histologic analyses, and functional drug screens (33). They found that each ALK inhibitor was associated with a distinct spectrum of ALK resistance mutations and that the frequency of one mutation—ALK G1202R—increased significantly after treatment with second-generation agents. Interestingly, they revealed that 33% of ALK+-tumors exhibited mutations within TP53, but it remained unclear whether these mutations were present prior therapy or occurred in the course of TKI-treatment. One year later, it was found by Aisner et al. that concurrent TP53 mutations are associated with a reduced survival in these patients (34), suggesting that molecular testing should be performed on all individuals with lung adenocarcinomas irrespective of clinical characteristics, in order to enable the detection of both targetable driver alterations and additional genetic alterations that have potential significance for therapy selection and as predictive markers for the efficacy of treatment. The authors assumed that TP53 mutations result in genetic instability and thereby trigger the development of multiple resistance mechanisms to targeted therapy reducing the survival rate in ALK+-patients. Just a few months later, this assumption could be confirmed by our group, by analyzing gene copy number alterations of 87 cancer-relevant genes in ALK+-tumors and cell lines harboring either wild-type or mutated TP53 with NanoString nCounter® technology (35). We found that TP53 mutations occurred in the early phase of tumorigenesis and that these mutations could lead to chromosomal instability. Our analyses further revealed that 24% of TP53-mutated patients had amplifications of the following cancer genes: MYC (14%), CCND1 (10%), TERT (5%), BIRC2 (5%), ORAOV1 (5%), YAP1 (5%). A summary of these findings is depicted in Figure 1.
By ChIP-Seq analyses we further found MYC-binding sites within the promoter region of EML4 in ALK+/TP53-mutated cells and MYC-overexpression resulted in elevated expression levels of the EML4-ALK protein, as well as increased cell proliferation rates, assuming a potential MYC-dependent resistance mechanism in patients with increased MYC copy numbers. In line with that and the aforementioned data by Aisner et al. (34), clinical data subsequently published by our group, confirmed that among 216 analyzed patients with ALK-rearranged NSCLC, the frequency of pathogenic TP53 mutations was 23.8%, while other co-occurring mutations were rare events in pre-treatment biopsies (36). It was further shown, that these patients had a significantly worse median progression-free survival (PFS) and overall survival (OS) compared to TP53 wild-type patients treated with either chemotherapy, crizotinib only or crizotinib followed by next-generation ALK-inhibitors [PFS 3.9 months (95% CI: 2.4–5.6) vs. 10.3 months (95% CI: 8.6–12.0), P<0.001; OS 15.0 months (95% CI: 5.0–24.9) vs. 50.0 months (95% CI: 22.9–77.1), P=0.002]. This difference was confirmed in all treatment-related subgroups, concluding that in ALK-rearranged NSCLC co-occurring TP53 mutations predict an unfavorable outcome of systemic therapies.
Taken together, these new findings show that the subgroup of ALK+-adenocarcinomas of the lung is more heterogeneous than historically assumed and will potentially have implications for stratification in future clinical trials for ALK+-patients. However, these studies are limited, as most lung cancer patients are diagnosed at advanced disease, when surgical resection is not possible. Therefore, diagnoses are performed on small biopsies, impeding comprehensive genomic analyses. It is of great importance to elucidate these genomic differences in more detail in the future in order to better understand the patient’s response to treatment and thereby improve their outcome.
The emergence of immunotherapy approaches, including monoclonal antibodies directed against cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) and programmed cell death protein-1 (PD-1) and its ligand (PD-L1) has redefined the management of lung cancer, achieving significant long-lasting responses with manageable safety profile. Current efforts are focusing on new potential combination strategies, using immune checkpoint blockade as a partner for targeted agents. However, there are few data available on the combination of checkpoint inhibitors with ALK TKIs in advanced NSCLC, and clinical trials are mostly still ongoing. Generally, it has been reported that patients with lung cancer caused by mutations or rearrangements within driver genes, such as ALK, do not show high response rates to immunotherapy in combination with checkpoint inhibitors as comprehensively reviewed in (37). In line with that, preclinical data on the treatment naïve ALK+-cell line H3122, which was administered to a combination of PD-1 and ALK inhibitor, did not show any synergistic tumor killing effects (38). It is known that tumor mutational burden (TMB), meaning the number of mutations carried by tumor cells, has an impact on tumor immunogenicity (39). TMB is reduced in lung cancers harboring ALK fusions as known drivers (40), which could explain the poor response to immunotherapy. A recent cohort study of 13 patients with advanced ALK+-NSCLC treated with a combination of nivolumab, an immune checkpoint inhibitor antibody which blocks PD-1, and crizotinib did not meet the primary endpoint of safety and tolerability for the first-line treatment, concluding that the findings do not support further evaluation of nivolumab and crizotinib in combination (41). In contrast, it was recently reported that the combination of full dose alectinib and atezolizumab in treatment-naïve ALK+-patients appeared to have an acceptable safety profile with no new safety findings for either agent. Despite the encouraging early efficacy results, further follow-up data are needed to define the benefit of this combination therapy (42). Retrospectively, it has to be noted that these patients were not stratified according to co-occurring mutations within other genes, such as TP53, which, as indicated by our own data, might be of great importance in future clinical settings.
Over the last years, our knowledge on ALK+-NSCLC has improved and revealed that the group of ALK+-NSCLC is more heterogeneous than initially thought. These new findings encourage intensive future research in order to better elucidate and understand the underlying molecular mechanisms occurring in drug resistance. This will lead to the development of new and more durable treatment strategies, which are even more personalized and consequently improve treatment outcome in patients.
Taken together, based on the current literature and our own findings, we suggest that an up-front assessment of TP53 status in all ALK+-patients should be implemented in the routine clinical work-up. A rapid and cost-effective pre-screening could be performed by means of p53 IHC, knowing that TP53 missense mutations result in a non-functional protein accumulation in the tumor cell nuclei, whereas truncating mutations result in the complete loss of p53. “Positive” cases should undergo additional sequencing analyses including assessment of copy number variations. In addition, it might be of importance to extend tumor analyses and include the determination of fusion partners and variants by using RNA-sequencing methods, such as Archer®FusionPlex® in the future to fully capture the genomic complexity of this group of genetically diverse tumors.
Acknowledgements
The group of Anne Maria Schultheis was funded by Roche Pharma AG and the Kölner Krebsstiftung.
Footnote
Conflicts of Interest: J Wolf declares advisory boards and lecture fees from Abbvie, AstraZeneca, BMS, Boehringer-Ingelheim, Chugai, Ignyta, Lilly, MSD, Novartis, Pfizer, Roche; research support (to institution) from BMS, MSD, Novartis, Pfizer. R Buettner is a founder and Chief Scientific Advisor for Targos, Mol Pathol, Kassel, Germany. The other authors have no conflicts of interest to declare.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25 Suppl 3:iii27-39. [Crossref] [PubMed]
- Boolell V, Alamgeer M, Watkins DN, et al. The Evolution of Therapies in Non-Small Cell Lung Cancer. Cancers (Basel) 2015;7:1815-46. [Crossref] [PubMed]
- Yu Y, Ding Z, Zhu L, et al. Frequencies of ALK rearrangements in lung adenocarcinoma subtypes: a study of 2299 Chinese cases. Springerplus 2016;5:894. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009;22:508-15. [Crossref] [PubMed]
- Inamura K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front Oncol 2017;7:193. [Crossref] [PubMed]
- Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene 1997;14:439-49. [Crossref] [PubMed]
- Solomon B, Wilner KD, Shaw AT. Current status of targeted therapy for anaplastic lymphoma kinase-rearranged non-small cell lung cancer. Clin Pharmacol Ther 2014;95:15-23. [Crossref] [PubMed]
- Murray PB, Lax I, Reshetnyak A, et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal 2015;8:ra6. [Crossref] [PubMed]
- Stoica GE, Kuo A, Aigner A, et al. Identification of anaplastic lymphoma kinase as a receptor for the growth factor pleiotrophin. J Biol Chem 2001;276:16772-9. [Crossref] [PubMed]
- Stoica GE, Kuo A, Powers C, et al. Midkine binds to anaplastic lymphoma kinase (ALK) and acts as a growth factor for different cell types. J Biol Chem 2002;277:35990-8. [Crossref] [PubMed]
- Bennasroune A, Mazot P, Boutterin MC, et al. Activation of the orphan receptor tyrosine kinase ALK by zinc. Biochem Biophys Res Commun 2010;398:702-6. [Crossref] [PubMed]
- Holla VR, Elamin YY, Bailey AM, et al. ALK: a tyrosine kinase target for cancer therapy. Cold Spring Harb Mol Case Stud 2017;3:a001115. [Crossref] [PubMed]
- Iyevleva AG, Raskin GA, Tiurin VI, et al. Novel ALK fusion partners in lung cancer. Cancer Lett 2015;362:116-21. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Sabir SR, Yeoh S, Jackson G, et al. EML4-ALK Variants: Biological and Molecular Properties, and the Implications for Patients. Cancers (Basel) 2017.9. [PubMed]
- Richards MW, O’Regan L, Roth D, et al. Microtubule association of EML proteins and the EML4-ALK variant 3 oncoprotein require an N-terminal trimerization domain. Biochem J 2015;467:529-36. [Crossref] [PubMed]
- Vendrell JA, Taviaux S, Beganton B, et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep 2017;7:12510. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Peters S, Adjei AA, Gridelli C, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2012;23 Suppl 7:vii56-64. [Crossref] [PubMed]
- Camidge DR, Doebele RC. Treating ALK-positive lung cancer--early successes and future challenges. Nat Rev Clin Oncol 2012;9:268-77. [Crossref] [PubMed]
- Doebele RC, Pilling AB, Aisner DL, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Lovly CM, McDonald NT, Chen H, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med 2014;20:1027-34. [Crossref] [PubMed]
- Kim HR, Kim WS, Choi YJ, et al. Epithelial-mesenchymal transition leads to crizotinib resistance in H2228 lung cancer cells with EML4-ALK translocation. Mol Oncol 2013;7:1093-102. [Crossref] [PubMed]
- Ji C, Zhang L, Cheng Y, et al. Induction of autophagy contributes to crizotinib resistance in ALK-positive lung cancer. Cancer Biol Ther 2014;15:570-7. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Markham A. Brigatinib: First Global Approval. Drugs 2017;77:1131-5. [Crossref] [PubMed]
- Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol 2017;18:1590-9. [Crossref] [PubMed]
- Recondo G, Facchinetti F, Olaussen KA, et al. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol 2018;15:694-708. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Aisner DL, Sholl LM, Berry L, et al. The Impact of Smoking and TP53 mutations in lung adenocarcinoma patients with targetable mutations-the Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 2018;24:1038-47. [Crossref] [PubMed]
- Alidousty C, Baar T, Martelotto LG, et al. Genetic instability and recurrent MYC amplification in ALK-translocated NSCLC: a central role of TP53 mutations. J Pathol 2018;246:67-76. [Crossref] [PubMed]
- Kron A, Alidousty C., et al. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol 2018;29:2068-75. [Crossref] [PubMed]
- Moya-Horno I, Viteri S, Karachaliou N, et al. Combination of immunotherapy with targeted therapies in advanced non-small cell lung cancer (NSCLC). Ther Adv Med Oncol 2018;10:1758834017745012. [Crossref] [PubMed]
- Hong S, Chen N, Fang W, et al. Upregulation of PD-L1 by EML4-ALK fusion protein mediates the immune escape in ALK positive NSCLC: Implication for optional anti-PD-1/PD-L1 immune therapy for ALK-TKIs sensitive and resistant NSCLC patients. Oncoimmunology 2015;5:e1094598. [Crossref] [PubMed]
- Pilotto S, Molina-Vila MA, Karachaliou N, et al. Integrating the molecular background of targeted therapy and immunotherapy in lung cancer: a way to explore the impact of mutational landscape on tumor immunogenicity. Transl Lung Cancer Res 2015;4:721-7. [PubMed]
- Spigel DR, Schrock AB, Fabrizio D. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol 2016;34:abstr 9017.
- Spigel DR, Reynolds C, Waterhouse D, et al. Phase 1/2 Study of the Safety and Tolerability of Nivolumab Plus Crizotinib for the First-Line Treatment of Anaplastic Lymphoma Kinase Translocation - Positive Advanced Non-Small Cell Lung Cancer (CheckMate 370). J Thorac Oncol 2018;13:682-8. [Crossref] [PubMed]
- Kim DW, Gadgeel SM, Gettinger SN, et al. Safety and clinical activity results from a phase Ib study of alectinib plus atezolizumab in ALK+ advanced NSCLC (aNSCLC). J Clin Oncol 2018;36:abstr 9009.


