
Are there any theranostic biomarkers in small cell lung carcinoma?
Introduction
Small cell lung cancer (SCLC) is an aggressive neuroendocrine neoplasia, strongly related to smoking attitude, characterized by rapid growth and very poor overall survival (1). It accounts for 13–15% of all lung cancers and represents the sixth common cause of death for malignant tumours (2).
Histological evaluation is mandatory. According to the World Health Organization (WHO), morphology and immunohistochemistry should be performed for the final diagnosis (3). Prognosis and management of patients are mainly related to tumour staging (4).
Most patients are diagnosed with advanced disease often with a metastatic dissemination [extensive stage (ES)]. Despite several research efforts, the survival remains poor and only slight improvements have been made for appropriate management and effective therapies. Based on the most recent ESMO guidelines, the management of the disease consists of chemotherapy and radiotherapy. A surgical approach is limited to a small percentage of localized cancers (T1-2, N0-1, M0) (1). The standard chemotherapy has not significantly changed in the last decades. Despite a good response after the first treatments, due to the elevated proliferative index and the sensitivity to DNA damaging drugs, the neoplasia acquires a chemo-resistance whose mechanisms remain unclear. Unlike NSCLC, a targeted therapy for SCLC has not yet been defined. The biology of SCLC is indeed not completely understood. Several studies have focused on the research of the molecular mechanisms responsible for the development, clinical behaviour and tumour landscape but the results are not conclusive (5). In the last five years some advances at the molecular level have resulted in the development of prognostic biomarkers and novel target agents (Figure 1).
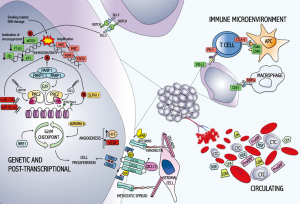
In this review we summarized some of the most important reports on biomarkers in the sections entitled: (I) cancer biomarkers (from genetic to post-transcriptional alterations); (II) immune microenvironment biomarkers; (III) circulating biomarkers.
Cancer biomarkers (from genetic to post-transcriptional alterations)
The molecular events which cause SCLC have not been clearly defined and a whole genetic characterization remains challenging. Only a few cases are treated by surgery and therefore tumour tissue available for molecular analysis is lacking. Recently, some efforts have been done to obtain more in depth information about the molecular signature of the neoplasia.
TP53 and Rb have been the most important molecular pathways investigated in SCLC. The inactivation of the two genes in a mouse model caused a high incidence of murine SCLC similar to the humans. This finding, together with the high penetrance of murine SCLC, was considered an important first step for testing new treatment modalities for SCLC (6).
More systematic genomic analyses were carried out several years later. In 2012 Peifer et al. conducted a wide analysis by using SNP array, exome sequencing, transcriptome and genome sequencing on a broad number of SCLC specimens. The results confirmed the inactivation of Rb and TP53, revealed mutations in PTEN, in SLIT2, and EPHA7, and focal amplifications of the MYC family and FGFR1. Furthermore, recurrent mutations in histone-modifying genes, CREBBP, EP300, and MLL were detected. These results increased the knowledge of the main biological events for the development of the neoplasia with the identification of new potential targetable genome alterations (7).
In the same year, Sos et al. performed a genomic and chemical vulnerability analysis on SCLC cell lines in order to identify therapeutically relevant genome alterations. The authors found that a subset of SCLC was susceptible to the action of Aurora B kinase inhibitors. This was another step to further investigation of the molecular basis in SCLC and consequently to a rational therapeutic approach (8).
A more in depth analysis on human tissue, SCLC and lymphoblastoid cell lines identified new recurrent somatic mutations. Among them, mutations in kinases, G-protein-coupled receptors, chromatin-modifying protein and SOX gene family members were detected. In addition, analysis of RNA sequencing data showed a recurrent RLF-MYCL1 fusion whose silencing obtained in cell lines decreased the proliferative activity (9).
Genome-wide copy number analysis found that amplifications of loci of MYC family genes were frequent and mutually exclusive. KIAA1432, was also identified as amplified in SCLC with frequent fusion transcripts in several amplified regions. This suggested a simultaneous occurrence of amplification and fusion of genes in a related way (10).
In 2015 the same group further explored the genes frequently mutated and expressed in SCLC potentially targetable in therapy. In addition to the most frequent TP53, Rb1 and PTEN alterations other genes, in particular TMEM132D, SPTA1 and VPS13B, were found, representing other therapeutic targets for the neoplasm (11).
In a relevant number of SCLC cases together with the main inactivation of TP53 and Rb, an inactivated mutation of NOTCH family genes was detected. This group of genes appeared involved in neuroendocrine differentiation in SCLC, as confirmed in a pre-clinical mouse model (12).
More recently the protein DLL-3 has been studied as a potential targetable biomarker in SCLC. This protein is normally expressed in the fetal brain where it acts on the somitogenesis and is an inhibitory ligand for the Notch pathway, suppressing oncogenesis and tumour growth.
In 2015 Saunders et al. discovered the association between DLL3 expression and the neuroendocrine phenotype. The study, carried out on xenograft models and human neoplastic tissue, demonstrated the up-regulation and the aberrant expression of DLL3 in SCLC. In this tumour, the employment of an anti-DLL3 treatment resulted effective in eradicating tumour initiating cells (13).
Based on these results several subsequent studies and clinical trials further investigated the role of DLL-3 and the efficacy of the administration of conjugated drugs for a target therapy in SCLC (14,15).
Some studies focused on the role of chromatin remodeling by chromatin modifying complex Polycomb Repressive Complex 2 (PRC2). This macromolecule is composed of subunits among which EZH1 or EZH2 (enhancer of zeste homolog), two histone H3K27 methyltransferases.
Already in 2013 the PRC2 overexpression and PRC2-target gene repression, such as cellular adhesion-related genes, was demonstrated to be associated with a poor prognosis in SCLC (16).
In the same year, Coe et al. studied the function of EZH2 as an oncogene. The high and aberrant expression of the histone methyltransferase was considered responsible for the hypermethylation of PRC2-target genes with consequent pro-tumourigenic functions in vitro (17).
The protein was discovered to work also on pro-apoptotic genes (Puma and Bad) whose inhibition promoted the proliferative activity and interfered with p21 levels (18). More recently findings from multiple patient-derived xenografts have linked the up-regulation of EZH2 with H3K27me3 associated SLFN11 gene silencing as a frequent mechanism of acquired chemoresistance in SCLC (19). Several EZH2 inhibitors are now under investigation for the treatment of different malignancies but not yet for SCLC.
An important role in SCLC biology has been demonstrated by non-coding RNA (ncRNA).
NcRNA are transcripts not translated into protein that are involved in key molecular processes (gene expression, genetic imprinting, histone modification, chromatin dynamics, etc.) through the interaction with all kinds of molecules. Several types of non-coding RNA are known. Among them, small non-coding RNA (sncRNA) and long non-coding RNA (lncRNA) are functionally important in SCLC. The distinction between the two groups is based on the number of nucleotides (less than and more than 200, respectively).
A lncRNA, CCAT2 (colon cancer-associated transcript 2), was detected in SCLC tissue and cell lines and its over-expression associated with advanced stage and poor prognosis (20). Recently the same group has identified a novel lncRNA, BLACAT1 (bladder cancer-associated transcript 1) involved in the biology of SCLC. BLACAT1 expression was higher in SCLC and was related to a suppressive activity in proliferation, migration and invasion, suggesting an oncogenic function. The authors concluded that the BLACAT1 acted as an oncogene correlated with a worse clinical status and outcome in SCLC patients (21).
Niu et al. have found that the expression of a splice variant of LIM-kinase-2 (LIMK2b) and its binding with EZH2 was regulated by the lncRNA TUG1 (taurine up-regulated gene 1). TUG1 was over-expressed in SCLC tissues promoting cell growth and chemo-resistance (22).
Similarly, the pathway of EZH1 protein was regulated by another lncRNA, HOTTIP (HOXA transcript at the distal tip) whose amplification in SCLC was associated with a more aggressive biological behavior (23).
In the group of ncRNA, miRNAs (microRNA) were also studied in SCLC.
Several miRNAs, such as miR-134 and miR-92a-2, were suggested to be involved in the mechanism of chemo-resistance by interfering in different processes. Some of these are the HOXA1 function, ZEB2 expression, regulation of apoptosis and autophagy, Kras/mapk pathway, PARP1 activity and epithelial mesenchymal transition (24-32).
A mechanism often damaged in the complex biology of SCLC is DNA repair. In this context, PARP1 [poly (ADP-ribose) polymerase 1] plays a fundamental role. The protein is involved in the repair of single-stranded DNA (ssDNA) and is found over-expressed in many gynecological cancers. Given the high level of DNA damage PARP protein levels are upregulated in SCLC more than other cancers, as demonstrated by a study of proteomic and gene expression profile (33).
Based on these data, the authors speculated on the usefulness of PARP1-inhibitors, evaluating the combination with other treatments. This inhibition acts on the E2F1 pathways, strengthening the therapeutic efficacy of inducing double-strand DNA break agents (33).
The efficacy of a PARP inhibitor BMN 673 was tested in vitro and in xenograft models by Cardnell et al. (34). Drug sensitivity was associated with high expression of DNA repair proteins while resistance was related to baseline activation of PI3K/mTOR pathway.
The association between PARP inhibitors and alkylating agents in SCLC has been reported recently. Interestingly, the expression of SLFN11 (schlafen family member 11), a guardian of the genome that promotes cell death in response to DNA damage, emerged as a strong predictor of the SCLC sensibility to the combination therapy of PARP inhibitors with temozolomide. Several clinical trials of a number of PARP inhibitors are ongoing in patients with SCLC (5,35).
Gardner et al. confirmed the link between the high levels of SLFN11 and the chemo-sensitivity of SCLC and calls into question the remodeling of chromatin EZH2-mediated as being responsible for silencing of SLFN11, re-establishing in vitro the levels of protein after EZH2-inhibition (19).
Another factor which emerged as a promising target for therapy in SCLC is WEE1, a kinase regulating cell cycle progression. The efficacy of WEE1 inhibitor (AZD1775) is limited by a common resistance. Sen et al. focused on the resistance mechanisms in order to enhance the treatment response. The authors found that the expression of AXL, a receptor tyrosine kinase, promoted the resistance by silencing mTOR cascade and activating an alternative DNA damage repair pathway (36).
Chemoresistance and early metastatic spread are the main factors for SCLC aggressive behavior. Here below are reported some of the most important key post-transcriptional molecular factors involved in metastatic spread.
The chemokine receptor CXCR4 was reported to be involved in SCLC progression in 2002 by Kijima et al. The authors found that all SCLC cell lines expressed CXCR4 receptors as well as c-kit even if in a lower percentage, suggesting their role in the pathogenesis of the neoplasm. The administration of small-molecule inhibitors demonstrated that the two pathways were cooperative in biological and biochemical functions and therefore the inhibition of both signals could be a targetable marker (37).
The high expression of CXCR4 was also confirmed in 2003 both on cell lines and primary tumour samples. Insights on its ligand CXCL12 and interaction with integrin signaling suggested a key role in cell motility and migration. This link gives resistance to chemotherapy thus its inhibition was reported as new therapeutic strategies for SCLC patients (38,39).
Further investigations demonstrated that this pathway was mainly mediated by a2, a4, a5, and b1 integrins whose activation resulted in an increased adhesion of SCLC cells to fibronectin and collagen. Stromal cells offer protection against chemotherapy-induced apoptosis thus the inhibition of this mechanism such as the T14 molecule and its derivatives and Plerixafor (AMD3100) could be helpful in overcoming the resistance (40,41).
Other works demonstrated alternative roles for CXCR4. In 2009 Pfeiffer et al. focused on the activation of the JAK2/STAT3 pathway by the link of CXCR4/CXCL12. The authors observed high levels of phosphorylated STAT3 in SCLC tissues, suggesting an important role in tumour biology. In cell lines STAT3 phosphorylation was demonstrated to be increased after CXCL12 stimulation through a cascade involving JAK2. These results supported an activity of CXCR4 both in VCAM1-mediated cell adhesion, spreading and cell growth through JAK2/STAT3 as mediators (42).
The inhibitor effect of the blockade of CXCR4 was investigated in an orthotopic xenograft mouse model. In mice with chemoresistant tumours, the administration of the molecule AMD3100 reduced tumour size and metastases formation, suggesting that the combination of CXCR4 inhibitors and standard chemotherapy could improve the neoplastic response and survival (43).
CXCR4 was confirmed to be expressed at high levels in SCLC and to correlate with bone and brain metastases, acting as an independent prognostic factor for survival (44).
The combination of the CXCR4 antagonist LY2510924 to chemotherapy was the object of a multicenter, open-label, randomized phase II study but the addition of this drug in extensive-disease SCLC did not seem to improve therapeutic efficacy (45).
High levels of CXCR4 and uPAR (urokinase receptor, a key factor expressed in renal tumours) were associated with increased lymph-nodal metastases and worse prognosis. In cell lines, their co-expression showed a higher invasive and migrating capacity. When injected in mice these were responsible for an increase in tumour size and the induction of lung metastases (46).
Overexpression of uPAR and HIF-1α/HIF-2α (transcription factors, involved in the angiogenetic process) were detected in tissues of SCLC patients with shorter overall survival (47). HIF-1α overexpression was more recently confirmed as an independent poor prognostic factor in SCLC patients (48).
Overexpression of VEGF, another important pathway involved in angiogenesis and metastatic processes, was found strictly related to high vascularization and nodal metastasis in ES-SCLC patients.
Nevertheless, at multivariate analysis, VEGF expression was the most important factor influencing overall survival. Higher expression of VEGF and survivin (inhibitor of apoptosis) was also detected in neoplastic tissue of SCLC patients compared to the control group and their expression was significantly associated with lymph node metastasis and shorter overall survival. Both factors resulted as independent poor prognostic factors (49).
Immune microenvironment biomarkers
Preclinical and clinical studies in the oncological field have demonstrated that the immune system has a crucial role in recognizing and eradicating tumour cells, thus providing a rationale for immunotherapy in different tumours (50).
In addition to its anticancer abilities, the immune system can also promote the growth and proliferation of tumour cells, since the microenvironment often contains immunosuppressive cell types such as Tregs, myeloid-derived suppressor cells and tumour-associated macrophages (51). Thus, the imbalance between immune-inhibitory and -stimulating mechanisms in the tumour microenvironment is crucial in determining tumour immune response and the rate of tumour progression (51).
The immune system has also been demonstrated to be involved in the pathophysiology of SCLC, particularly in relation to the frequent occurrence of paraneoplastic disorders (e.g., Lambert-Eaton myasthenic syndrome) and the positive predictive impact of immune activity on patient prognosis (50).
Immune checkpoint pathways involving the cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein-1 (PD-1) receptors and their ligands dampen the T-cell immune response (52) and represent important biomarkers in different tumour types both for prognostic and therapeutic purposes. Nevertheless at present limited data have been reported on immune checkpoint blockade in patients with SCLC and only a few studies were investigated the prognostic and predictive role of this marker.
CTLA-4 is a transmembrane protein receptor expressed on the surface of T cells which regulate responses in the early stages of T-cell activation (53). CTLA-4 outcompetes the CD28 receptor for binding to its ligands, CD80/CD86, expressed by antigene-presenting cells (APCs), providing an inhibitory rather than stimulatory signal to the T cell. Thus, CTLA-4 is involved in the limitation of inflammatory responses and the tolerance to self-antigens, preventing autoimmunity. However, CTLA-4 may also play a detrimental role by inhibiting antitumor immunity and its blockade, therefore, may remove the inhibitory signal and stimulate antitumor immunity. It was the first immune checkpoint receptor to be targeted by a therapeutic agent (Ipilimumab) and it was tested in some trials, with positive results for progression-free survival but not for overall survival (5).
PD-1 is another important immune checkpoint receptor largely studied in oncology (54). It belongs to the CD28 family protein and it is expressed on the surface of T cells, regulating their activation and proliferation. It has been widely demonstrated that binding of PD-1 (expressed on activated tumor-infiltrating T cells) to PD-L1 (expressed on tumor cells and on tumor immune microenvironment) can inhibit antitumor immune responses in different malignancies, such as NSCLC, melanoma and renal cell cancer (55,56). In 2016, a multicentre, open-label, phase I/II trial of nivolumab (monoclonal antibody against PD-1) with or without ipilimumab in patients with recurrent ES-SCLC showed a better 1-year survival (57) leading to the incorporation of the nivolumab and ipilimumab combination as a National Comprehensive Cancer Network (NCCN) guideline recommendation for the second-line treatment of ES-SCLC. However, the efficacy of checkpoint inhibitors in patients with SCLC needs to be confirmed in randomized trials, and these treatments have not been formally approved for the treatment of SCLC (5). Only a few studies have investigated the distribution of PD-L1 expression in SCLC tissue samples, reporting contradictory data concerning immunostaining and correlation with clinical outcomes.
A paper exploring the PD-1/PD-L1 expression pattern in SCLC was published in 2015 by Schultheis et al., and none of the patients showed positive PD-L1 immunostaining in the tumour cell component, while 18.5% of them showed positivity in tumour-infiltrating macrophages and 48% in T-lymphocytes (58).
Other interesting studies showed a correlation between positive PD-L1 immunostaining and a longer overall survival (59) or disease-free survival (60). A comparative study using the three different antibodies was done by Takada et al. and reproducible data were found only when a 5% cut-off of PD-L1 positive immunostaining was chosen (61).
The predictive role of PD-L1 expression on tumour and different immune cell components was investigated by two studies that focused on brain metastases of SCLC and a positive prognostic role was found for PD-L1 expression, in particular in T-lymphocytes, supporting the hypothesis that an active immune microenvironment may be targettable in metastatic SCLC (62,63).
In 2016, for the first time, copy number alteration of CD274, the gene encoding PD-L1, was examined to determine in detail genomic alterations that may affect PD-L1 expression levels in SCLC. The authors reported that a subset (2%) of SCLC showed amplification of CD272, resulting in high immunohistochemical expression of PD-L1, concluding that the amplification of this region could be particularly sensitive to therapeutic PD-1/PD-L1 (64). In 2017 the first and only article about a negative prognostic impact of PD-L1 expression in SCLC was published by Chang et al. A direct correlation was found between tumour cells an T-lymphocyte PD-L1 expression, as well as with advanced disease stage (IV); multivariate analysis revealed that high tumour PD-L1 expression and stage IV disease represented two independent risk factors for poor overall survival (65).
Interestingly, the role of immune microenvironment in SCLC focusing on PD-L1 expression and FOXP3-positive tumour infiltrating T-lymphocytes was investigated in a large number of non-metastatic and metastatic SCLC tests. PD-L1 was more frequently detected in non-metastatic SCLC cases, leading to the hypothesis that downregulation of PD-L1 could be related to a high invasion potential. Another intriguing result was the potential prognostic impact of FOXP3+T-lymphocytes in the different stages of SCLC (66). More studies are needed to clarify the role of PD-L1 in SCLC, in terms of prevalence of PD-L1 expression and in evaluating its role in predicting response to the treatment and outcome.
Overexpression of CD47, a cell-surface molecule that inhibits activation and phagocytic activity of macrophages, was found on the surface of human SCLC cells and has been implicated in immune escape by tumours (67). To date, only experimental studies have been performed, and clinical exploration of CD47/SIRPα inhibition as a therapeutic strategy for SCLC is expected to begin.
Circulating biomarkers
Several efforts have been made for years to find cost-effective and non-invasive serum markers able to indicate the biology and the behavior of SCLC. Several serum biomarkers have been identified and reported in publications. However despite extensive studies, cancer detection and follow-up via a single marker remains difficult due to the low sensitivity, specificity and reproducibility of serum biomarkers. Multiple biomarkers rather than a single serum marker testing could be the most sensitive approach in the future.
The significance of neuron specific enolase (NSE) in SCLC has been investigated and reported to have a role in the diagnosis, treatment and follow-up since the mid-eighties. The levels of NSE were found to have a stronger correlation to disease extent and response to treatment in patients in comparison to other markers [such as carcinoembryonic antigen (CEA); calcitonin, ferritin, lactate dehydrogenase (LDH)] (68,69). The sensitivity and specificity of NSE serum marker has been addressed by several authors often with contradictory results. NSE levels in SCLC patients were considered to have a good specificity, whereas the sensitivity appeared depending on the stage of disease. Indeed, the levels were elevated in a majority of ES-SCLC and in a less percentage of limited one (70).
NSE measurements before therapy also resulted useful in evaluating response duration, as emerged from a multivariate analysis (71).
More recently, a meta-analysis about the association of serum NSE levels and the prognosis of SCLC confirmed that patients with high levels of NSE showed a shorter overall survival than patients with low ones, attributing a prognostic value to the marker (72).
In patients treated with first line platinum-based chemotherapy high levels of serum NSE as well as lactate dehydrogenase (LDH) had a worse progression free survival and overall survival, confirming a suggested prognostic and predictive function (73).
A high level of serum LDH, a metabolic enzyme detected in patients with several types of malignancies, was identified as a prognostic factor to better stratify patients with SCLC (74). Several studies found a significant relation between serum LDH levels and outcome either in limited-stage and extensive disease SCLC (75,76).
A potential validation for the use of LDH in monitoring treatment response and for discriminating different forms of diseases has also recently been proposed by Chen et al. (77).
Serum pro-gastrin-releasing peptide (ProGRP), a neuropeptide involved in several physiological mechanisms, was reported as a useful marker for treatment monitoring and survival in SCLC (78).
However the prognostic and predictive value of ProGRP remained doubtful as emerged from the results of multivariate analyses especially if compared to other markers such as NSE (79,80).
Higher levels of carcinoembryonic antigen (CEA), an oncofetal protein, in patients with ES-SCLC and a strong correlation with response and survival was firstly reported by Sculier et al. in 1985 (81).
The relevance of CEA serum concentrations in SCLC patients in predicting the outcome was suggested by Zhu et al. In extensive-stage disease, normal serum CEA levels together with thoracic irradiation therapy and more than 4 cycles of chemotherapy were independent prognostic factors for overall survival in SCLC (82).
CEA has also been recently investigated as predictor of brain metastasis and survival in prophylactic cranial irradiated SCLC patients (83).
CTCs (circulating tumour cells) are cancer cells that are shed into vessels from tumors and have the potential to develop metastatic lesions. SCLC is the cancer with the higher level of CTCs. Given the metastatic potential of these cells, many studies have been carried out to understand their diagnostic, prognostic and predictive value in neoplastic diseases and their utility as biomarker or as “liquid biopsy”. The prognostic value of CTCs in SCLC was controversial until the early 2000s mainly due not yet well developed technologies for detection (84).
In 2012, Hou et al. found that the CTCs, apoptotic CTCs and CTM (circulating tumor microemboli) number after one cycle of chemotherapy were independent prognostic factors for SCLC (85).
Several other groups confirmed more recently the link between CTCs and poor prognosis in SCLC patients (86-88). However the role of CTCs as predictors to therapeutic response remains quite debated. Wang et al. found relation between the CTCs levels and serum NSE but not with the therapy responsiveness (89).
The decrease in the number of CTCs in response to a second-line chemotherapy was proposed to be related to the effect on the metastases or resident SCLC lesions rather than an effective tumor progression blockage (90).
Several studies deepened the molecular biology of CTCs and highlighted different genetic profiles responsible of chemosensitive or chemorefractory SCLC (91,92). Several works have recently focused on the expression of different CTC biomarkers, such as receptor of tyrosine kinase, CXCR4, VEGFR2, Bcl-2, etc. and their correlation with prognosis and chemo-resistance mechanisms (93-95).
The CTCs are the responsible of the metastatic dissemination of SCLC. This is proven by the expression of epithelial-mesenchymal transition (EMT) phenotype in SCLC cell lines derived from primary tumors and metastases. The presence of a CTC with a mesenchymal-like phenotype, based on the low expression of E-cadherine and the high expression of c-MET, was associated with longer survival (96).
Acknowledgements
The authors thank Dr. Judith Wilson for English revision.
Footnote
Conflict of Interest: The authors have no conflicts of interest to declare.
References
- Früh M, De Ruysscher D, Popat S, et al. Small-cell lung cancer (SCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24 Suppl 6:vi99-105. [Crossref] [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2010, National Cancer Institute. Bethesda, MD, based on November 2012 SEER data submission, posted to the SEER web site, April 2013.
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Nicholson AG, Chansky K, Crowley J, et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the Revision of the Clinical and Pathologic Staging of Small Cell Lung Cancer in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:300-11.
- Sabari JK, Lok BH, Laird JH, et al. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 2017;14:549-61. [Crossref] [PubMed]
- Meuwissen R, Linn SC, Linnoila RI, et al. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003;4:181-9. [Crossref] [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Sos ML, Dietlein F, Peifer M, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci 2012;109:17034-9. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Iwakawa R, Takenaka M, Kohno T, et al. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosomes Cancer 2013;52:802-16. [Crossref] [PubMed]
- Iwakawa R, Kohno T, Totoki Y, et al. Expression and clinical significance of genes frequently mutated in small cell lung cancers defined by whole exome/RNA sequencing. Carcinogenesis 2015;36:616-21. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Saunders LR, Bankovich AJ, Anderson WC, et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci Transl Med 2015;7:302ra136. [Crossref] [PubMed]
- Bauer TM, Spigel D, Ready N, et al. ORAL02.01: Safety and Efficacy of Single-Agent Rovalpituzumab Tesirine, a DLL3-Targeted ADC, in Recurrent or Refractory SCLC: Topic: Medical Oncology. J Thorac Oncol 2016;11:S252-3. [Crossref] [PubMed]
- Lashari BH, Vallatharasu Y, Kolandra L, et al. Rovalpituzumab Tesirine: A Novel DLL3-Targeting Antibody-Drug Conjugate. Drugs R D 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Sato T, Kaneda A, Tsuji S, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep 2013;3:1911. [Crossref] [PubMed]
- Coe BP, Thu KL, Aviel-Ronen S, et al. Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer. PLoS One 2013;8:e71670. [Crossref] [PubMed]
- Hubaux R, Thu KL, Coe BP, et al. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 2013;8:1102-6. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Chen S, Wu H, Lv N, et al. LncRNA CCAT2 predicts poor prognosis and regulates growth and metastasis in small cell lung cancer. Biomed Pharmacother 2016;82:583-8. [Crossref] [PubMed]
- Chen W, Hang Y, Xu W, et al. BLACAT1 predicts poor prognosis and serves as oncogenic lncRNA in small-cell lung cancer. J Cell Biochem 2018. [Epub ahead of print]. [PubMed]
- Niu Y, Ma F, Huang W, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 2017;16:5. [Crossref] [PubMed]
- Sun Y, Zhou Y, Bai Y, et al. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol Cancer 2017;16:162. [Crossref] [PubMed]
- Guo L, Liu Y, Bai Y, et al. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer 2010;46:1692-702. [Crossref] [PubMed]
- Ranade AR, Cherba D, Sridhar S, et al. MicroRNA 92a-2*: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. J Thorac Oncol 2010;5:1273-8. [Crossref] [PubMed]
- Xiao F, Bai Y, Chen Z, et al. Downregulation of HOXA1 gene affects small cell lung cancer cell survival and chemoresistance under the regulation of miR-100. Eur J Cancer 2014;50:1541-54. [Crossref] [PubMed]
- Fang S, Zeng X, Zhu W, et al. Zinc finger E-box-binding homeobox 2 (ZEB2) regulated by miR-200b contributes to multi-drug resistance of small cell lung cancer. Exp Mol Pathol 2014;96:438-44. [Crossref] [PubMed]
- Bai Y, Sun Y, Peng J, et al. Overexpression of secretagogin inhibits cell apoptosis and induces chemoresistance in small cell lung cancer under the regulation of miR-494. Oncotarget 2014;5:7760-75. [Crossref] [PubMed]
- Yang X, Bai F, Xu Y, et al. Intensified Beclin-1 Mediated by Low Expression of Mir-30a-5p Promotes Chemoresistance in Human Small Cell Lung Cancer. Cell Physiol Biochem 2017;43:1126-39. [Crossref] [PubMed]
- Liu H, Huang J, Peng J, et al. Upregulation of the inwardly rectifying potassium channel Kir2.1 (KCNJ2) modulates multidrug resistance of small-cell lung cancer under the regulation of miR-7 and the Ras/MAPK pathway. Mol Cancer 2015;14:59. [Crossref] [PubMed]
- Luo Y, Tong L, Meng H, et al. MiR-335 regulates the chemo-radioresistance of small cell lung cancer cells by targeting PARP-1. Gene 2017;600:9-15. [Crossref] [PubMed]
- Wei T, Zhu W, Fang S, et al. miR-495 promotes the chemoresistance of SCLC through the epithelial-mesenchymal transition via Etk/BMX. Am J Cancer Res 2017;7:628-46. [PubMed]
- Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2:798-811. [Crossref] [PubMed]
- Cardnell RJ, Feng Y, Diao L, et al. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res 2013;19:6322-8. [Crossref] [PubMed]
- Lok BH, Gardner EE, Schneeberger VE, et al. PARP Inhibitor Activity Correlates with SLFN11 Expression and Demonstrates Synergy with Temozolomide in Small Cell Lung Cancer. Clin Cancer Res 2017;23:523-35. [Crossref] [PubMed]
- Sen T, Tong P, Diao L, et al. Targeting AXL and mTOR Pathway Overcomes Primary and Acquired Resistance to WEE1 Inhibition in Small-Cell Lung Cancer. Clin Cancer Res 2017;23:6239-53. [Crossref] [PubMed]
- Kijima T, Maulik G, Ma PC, et al. Regulation of cellular proliferation, cytoskeletal function, and signal transduction through CXCR4 and c-Kit in small cell lung cancer cells. Cancer Res 2002;62:6304-11. [PubMed]
- Burger M, Glodek A, Hartmann T, et al. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene 2003;22:8093-101. [Crossref] [PubMed]
- Hartmann TN, Burger M, Burger JA. The role of adhesion molecules and chemokine receptor CXCR4 (CD184) in small cell lung cancer. J Biol Regul Homeost Agents 2004;18:126-30. [PubMed]
- Hartmann TN, Burger JA, Glodek A, et al. CXCR4 chemokine receptor and integrin signaling co-operate in mediating adhesion and chemoresistance in small cell lung cancer (SCLC) cells. Oncogene 2005;24:4462-71. [Crossref] [PubMed]
- Burger JA, Stewart DJ. CXCR4 chemokine receptor antagonists: perspectives in SCLC. Expert Opin Investig Drugs 2009;18:481-90. [Crossref] [PubMed]
- Pfeiffer M, Hartmann TN, Leick M, et al. Alternative implication of CXCR4 in JAK2/STAT3 activation in small cell lung cancer. Br J Cancer 2009;100:1949-56. [Crossref] [PubMed]
- Taromi S, Kayser G, Catusse J, et al. CXCR4 antagonists suppress small cell lung cancer progression. Oncotarget 2016;7:85185-95. [Crossref] [PubMed]
- Li XX, Li RJ, Zhao LJ, et al. Expression of molecular factors correlated with metastasis in small cell lung cancer and their significance. Int J Clin Exp Pathol 2015;8:14676-84. [PubMed]
- Salgia R, Stille JR, Weaver RW, et al. A randomized phase II study of LY2510924 and carboplatin/etoposide versus carboplatin/etoposide in extensive-disease small cell lung cancer. Lung Cancer 2017;105:7-13. [Crossref] [PubMed]
- Li Y, Shen Y, Miao Y, et al. Co -expression of uPAR and CXCR4 promotes tumor growth and metastasis in small cell lung cancer. Int J Clin Exp Pathol 2014;7:3771-80. [PubMed]
- Yang SL, Ren QG, Wen L, et al. Clinicopathological and prognostic significance of hypoxia-inducible factor-1 alpha in lung cancer: a systematic review with meta-analysis. J Huazhong Univ Sci Technolog Med Sci 2016;36:321-7. [Crossref] [PubMed]
- Lin CS, Liu TC, Lee MT, et al. Independent Prognostic Value of Hypoxia-inducible Factor 1-alpha Expression in Small Cell Lung Cancer. Int J Med Sci 2017;14:785-90. [Crossref] [PubMed]
- Chen P, Zhu J, Liu DY, et al. Over-expression of survivin and VEGF in small-cell lung cancer may predict the poorer prognosis. Med Oncol 2014;31:775. [Crossref] [PubMed]
- Horn L, Reck M, Spigel DR. The Future of Immunotherapy in the Treatment of Small Cell Lung Cancer. Oncologist 2016;21:910-21. [Crossref] [PubMed]
- Reck M, Heigener D, Reinmuth N. Immunotherapy for small-cell lung cancer: emerging evidence. Future Oncol 2016;12:931-43. [Crossref] [PubMed]
- Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol 2006;90:51-81. Review. [Crossref] [PubMed]
- Wolchok JD, Saenger Y. The mechanism of anti-CTLA-4 activity and the negative regulation of T-cell activation. Oncologist 2008;13 Suppl 4:2-9. [Crossref] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252-64. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [Crossref] [PubMed]
- Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. [Crossref] [PubMed]
- Schultheis AM, Scheel AH, Ozretić L, et al. PD-L1 expression in small cell neuroendocrine carcinomas. Eur J Cancer 2015;51:421-6. [Crossref] [PubMed]
- Ishii H, Azuma K, Kawahara A, et al. Significance of programmed cell death-ligand 1 expression and its association with survival in patients with small cell lung cancer. J Thorac Oncol 2015;10:426-30. [Crossref] [PubMed]
- Toyokawa G, Takada K, Haratake N, et al. Favorable Disease-free Survival Associated with Programmed Death Ligand 1 Expression in Patients with Surgically Resected Small-cell Lung Cancer. Anticancer Res 2016;36:4329-36. [PubMed]
- Takada K, Toyokawa G, Okamoto T, et al. An Immunohistochemical Analysis of PD-L1 Protein Expression in Surgically Resected Small Cell Lung Cancer Using Different Antibodies and Criteria. Anticancer Res 2016;36:3409-12. [PubMed]
- Berghoff AS, Ricken G, Wilhelm D, et al. Tumor infiltrating lymphocytes and PD-L1 expression in brain metastases of small cell lung cancer (SCLC). J Neurooncol 2016;130:19-29. [Crossref] [PubMed]
- Liu J, Lu Z, Wang W, et al. Programmed death-ligand 1 positivity can predict improved survival and a lower risk of brain metastasis in patients with resectable small cell lung cancer. Oncol Lett 2018;16:2373-81. [PubMed]
- George J, Saito M, Tsuta K, et al. Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer. Clin Cancer Res 2017;23:1220-6. [Crossref] [PubMed]
- Chang YL, Yang CY, Huang YL, et al. High PD-L1 expression is associated with stage IV disease and poorer overall survival in 186 cases of small cell lung cancers. Oncotarget 2017;8:18021-30. [PubMed]
- Bonanno L, Pavan A, Dieci MV, et al. The role of immune microenvironment in small-cell lung cancer: Distribution of PD-L1 expression and prognostic role of FOXP3-positive tumour infiltrating lymphocytes. Eur J Cancer 2018;101:191-200. [Crossref] [PubMed]
- Weiskopf K, Jahchan NS, Schnorr PJ, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest 2016;126:2610-20. [Crossref] [PubMed]
- Fischbach W, Jany B. Neuron-specific enolase in the diagnosis and therapy monitoring of lung cancer: a comparison with CEA, TPA, ferritin and calcitonin. Int J Biol Markers 1986;1:129-36. [Crossref] [PubMed]
- Jørgensen LG, Hansen HH, Cooper EH. Neuron specific enolase, carcinoembryonic antigen and lactate dehydrogenase as indicators of disease activity in small cell lung cancer. Eur J Cancer Clin Oncol 1989;25:123-8. [Crossref] [PubMed]
- Burghuber OC, Worofka B, Schernthaner G, et al. Serum neuron-specific enolase is a useful tumor marker for small cell lung cancer. Cancer 1990;65:1386-90. [Crossref] [PubMed]
- Jørgensen LG, Osterlind K, Hansen HH, et al. Serum neuron specific enolase (NSE) is a determinant of response duration in small cell lung cancer (SCLC). Br J Cancer 1992;66:594-8. [Crossref] [PubMed]
- Zhao WX, Luo JF. Serum neuron-specific enolase levels were associated with the prognosis of small cell lung cancer: a meta-analysis. Tumour Biol 2013;34:3245-8. [Crossref] [PubMed]
- Liu X, Zhang W, Yin W, et al. The prognostic value of the serum neuron specific enolase and lactate dehydrogenase in small cell lung cancer patients receiving first-line platinum-based chemotherapy. Medicine (Baltimore) 2017;96:e8258. [Crossref] [PubMed]
- Stokkel MP, van Eck-Smit BL, Zwinderman AH, et al. Pretreatment serum LDH as additional staging parameter in small-cell lung carcinoma. Neth J Med 1998;52:65-70. [Crossref] [PubMed]
- Tas F, Aydiner A, Demir C, et al. Serum lactate dehydrogenase levels at presentation predict outcome of patients with limited-stage small-cell lung cancer. Am J Clin Oncol 2001;24:376-8. [Crossref] [PubMed]
- Hermes A, Gatzemeier U, Waschki B, et al. Lactate dehydrogenase as prognostic factor in limited and extensive disease stage small cell lung cancer – a retrospective single institution analysis. Respir Med 2010;104:1937-42. [Crossref] [PubMed]
- Chen C, Zhu YH, Huang JA. Clinical evaluation of potential usefulness of serum lactate dehydrogenase level in follow-up of small cell lung cancer. J Cancer Res Ther 2018;14:S336-40. [Crossref] [PubMed]
- Sunaga N, Tsuchiya S, Minato K, et al. Serum pro-gastrin-releasing peptide is a useful marker for treatment monitoring and survival in small-cell lung cancer. Oncology 1999;57:143-8. [Crossref] [PubMed]
- Huang Z, Xu D, Zhang F, et al. Pro-gastrin-releasing peptide and neuron-specific enolase: useful predictors of response to chemotherapy and survival in patients with small cell lung cancer. Clin Transl Oncol 2016;18:1019-25. [Crossref] [PubMed]
- Cavalieri S, Morelli D, Martinetti A, et al. Clinical implications for pro-GRP in small cell lung cancer. A single center experience. Int J Biol Markers 2018;33:55-61. [Crossref] [PubMed]
- Sculier JP, Feld R, Evans WK, et al. Carcinoembryonic antigen: a useful prognostic marker in small-cell lung cancer. J Clin Oncol 1985;3:1349-54. [Crossref] [PubMed]
- Zhu H, Guo H, Li M, et al. Increased serum carcinoembryonic antigen level can predict poor survival of patients with small cell lung cancer. Transl Res 2015;166:355-65. [Crossref] [PubMed]
- Guo D, Jing W, Zhu H, et al. Clinical value of carcinoembryonic antigen for predicting the incidence of brain metastases and survival in small cell lung cancer patients treated with prophylactic cranial irradiation. Cancer Manag Res 2018;10:3199-205. [Crossref] [PubMed]
- Ma PC, Blaszkowsky L, Bharti A, et al. Circulating tumor cells and serum tumor biomarkers in small cell lung cancer. Anticancer Res 2003;23:49-62. [PubMed]
- Hou JM, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525-32. [Crossref] [PubMed]
- Naito T, Tanaka F, Ono A, et al. Prognostic impact of circulating tumor cells in patients with small cell lung cancer. J Thorac Oncol 2012;7:512-9. [Crossref] [PubMed]
- Hiltermann TJ, Pore MM, van den Berg A, et al. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol 2012;23:2937-42. [Crossref] [PubMed]
- Normanno N, Rossi A, Morabito A, et al. Prognostic value of circulating tumor cells' reduction in patients with extensive small-cell lung cancer. Lung Cancer 2014;85:314-9. [Crossref] [PubMed]
- Wang X, Ma K, Wang Y, et al. Evaluation of Circulating Tumor Cells in Predicting Therapeutic Response in Small Cell Lung Cancer Patients. Arch Med Res 2016;47:454-9. [Crossref] [PubMed]
- Hamilton G, Rath B, Holzer S, et al. Second-line therapy for small cell lung cancer: exploring the potential role of circulating tumor cells. Transl Lung Cancer Res 2016;5:71-7. [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med 2017;23:114-9. [Crossref] [PubMed]
- Hamilton G, Rath B, Klameth L, et al. Receptor tyrosine kinase expression of circulating tumor cells in small cell lung cancer. Oncoscience 2015;2:629-34. [Crossref] [PubMed]
- Messaritakis I, Politaki E, Kotsakis A, et al. Phenotypic characterization of circulating tumor cells in the peripheral blood of patients with small cell lung cancer. PLoS One 2017;12:e0181211. [Crossref] [PubMed]
- Messaritakis I, Nikolaou M, Politaki E, et al. Bcl-2 expression in circulating tumor cells (CTCs) of patients with small cell lung cancer (SCLC) receiving front-line treatment. Lung Cancer 2018;124:270-8. [Crossref] [PubMed]
- Pore M, Meijer C, de Bock GH, et al. Cancer Stem Cells, Epithelial to Mesenchymal Markers, and Circulating Tumor Cells in Small Cell Lung Cancer. Clin Lung Cancer 2016;17:535-42. [Crossref] [PubMed]


