
The microbiome and lung cancer
Human microbiome and cancer
The microbiome is the ecological community of commensal, symbiotic, and pathogenic microorganisms that share our body space (1). These microorganisms include protozoa, fungi, bacteria, and viruses, and form organ-specific microbial communities. The size and composition of the microbiome vary from one body part to another, are affected by host and environmental factors, and can contribute to both disease and the body’s response to it. According to current estimates, the number of microbial cells is equal to the number of host cells and the total microbiome constitutes 0.2 kg in weight (2). It is a well-known fact that microbial organisms have been the cause of infectious diseases and morbidity in humans. However, an increasing level of evidence currently supports the role of microbial etiopathogenesis for non-infectious diseases including cancer (3). Epidemiological studies in humans were initially able to show association of microorganisms with cancer. Animal models were then used to prove an etiopathogenic role of specific microorganisms in human cancer. The goal of this manuscript is to briefly summarize the current state of knowledge of various roles played by microorganisms as related to cancer. Additionally, the putative role of microbiome in lung cancer is discussed with a brief discussion on the methods for isolation of microbiome from human samples for genomic sequencing. The intent of the review is to provide a primer and a summary of current knowledge of the field to clinicians treating lung cancer.
Some microbes, especially viruses, are known to cause cancer. Since the early work on the oncogenic potential of Rous sarcoma virus (4), literature is replete with examples of such associations. Viruses that have been known to cause cancer in humans include human herpes viruses (HHV) 4 and 8, hepatitis B virus (HBV) and hepatitis C virus (HCV), human T-lymphotropic virus-I (HTLV-I), human papilloma viruses (HPV) 16 and 18, and Merkel cell polyomavirus (MCV) (5). Among bacteria, Helicobacter pylori (H. pylori) and Salmonella typhi bacteria are known to contribute to the development of gastric cancer and cholangiocarcinoma (6,7). The protozoal parasites Schistosoma haematobium and Opisthorchis viverrini may have a role in urinary bladder and gall bladder carcinogenesis (8). While it is documented that certain parasites, bacteria, and viruses carry oncogenic potential, it is not evident how the human microbiome affects cancer causation and prognosis. This body of literature is still evolving and has been made possible in large part by the recent technological advances such as high-throughput DNA sequencing.
Animal studies on association of microbiome with cancer
Previous studies using animal models have shown evidence for association of bacterial microbiota with tumors of gut, liver, kidney, breast, and lung (Table 1). Some of the pioneer animal studies utilized germ-free mouse and germ-free rat models to test whether microorganisms had any association with carcinogenesis. In a study of the germ-free rat model it was observed that although spontaneous tumors occurred and were similar to those in rats raised conventionally, the occurrence of solid tumors was significantly lower (9). In yet another study, the carcinogenicity of methylazoxymethanol-β-D-glucosiduronic acid was tested in conventional and germ-free rats (11). While the conventional rats developed tumors in the colon, kidney, and liver, the germ-free rats did not develop any tumor whether the carcinogen was given by oral or intraperitoneal routes (11). Similarly, another study showed that only 20% germ-free rats developed 1,2-dimethylhydrazine-induced colonic tumors, whereas 93% of conventional rats developed multiple colonic tumors (18). Other more recent studies investigated the mechanistic role of bacteria and bacterial induced inflammation in colon cancers. One of the studies explored the contribution of the host intestinal microbiota and inflammatory response, as a measure of MYD88 signaling, to the development of colitis-associated cancer. In this study conventional mice developed ulcerative colitis and subsequently colon carcinomas while germ-free mice remained tumor-free (12). The authors concluded that severity of chronic colitis directly correlated to colorectal tumor development and that bacterial-induced inflammation was responsible for progression from adenoma to invasive carcinoma (12). It was also found that rats that did not develop cancer had better anti-cancer immune response with an increase in B, cytolytic T, natural killer (NK), and NK T cells, and cytotoxicity in peripheral blood (13). This further indicates that the lower antigenic challenge and the absence of the physiological inflammation allows the germ-free rats to develop more efficacious anti-cancer immune responses (13). Overall, germ-free rat/mice models demonstrate the role of a putative pathogenic microbiome and inflammation leading to cancer. However, when it comes to the role of a single microorganism in cancer, the prominent example is that of H. pylori in gastric cancer. H. pylori is recognized as the major cause of gastric cancer and has been classified by World Health Organization as a group I carcinogen (19). H. pylori infection causes persistent chronic gastritis which, in susceptible individuals, may progress to intestinal-type gastric cancer. In studies of gastric cancer, H. pylori infection is known to progress over decades through stages of chronic gastritis, atrophy, intestinal metaplasia, dysplasia, and cancer. The development of gastric cancer has been attributed to alterations in DNA resulting from chronic inflammation, imbalance between epithelial-cell proliferation and apoptosis, and gastric colonization by enteric bacteria with nitrate reductase activity facilitating the formation of carcinogenic nitrosamines in an environment of gastric atrophy. The various stages of gastric cancer have been reproduced in mice (19). Additionally, it has been shown that the H. pylori virulence factors may have a direct role in gastric atrophy leading to gastric cancer (20). Importantly, eradication of H. pylori by early antibiotic therapy significantly reduced incidence of gastric cancer and delayed antibiotic therapy reduced the number of dysplastic lesions in transgenic insulin-gastrin (INS-GAS) mice (14). In a monospecies infection with H. pylori, germ-free INS-GAS mice developed gastrointestinal neoplasia, although it took 13 months longer than conventional mice, further confirming a role for intestinal microflora in progression of gastric cancer (21).
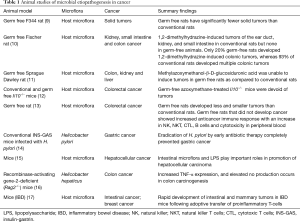
Full table
Liver cancer or hepatocellular carcinoma is preceded by chronic inflammation and fibrosis (22). Chronic liver diseases are associated with an increased translocation of intestinal bacteria and incidence of bacterial infections in patients (22). Concurrent to these findings, 40% of cirrhotic rats with ascites and 80% of cirrhotic rats with spontaneous bacterial peritonitis displayed bacterial translocation into their mesenteric lymph nodes (23). Additionally, chronic hepaptitis and associated liver cancer was promoted in Helicobacter hepaticus-infected mice (24). In a study performed in mice, the intestinal microflora as the main source for portal lipopolysaccharide (LPS) has been found to be an important prerequisite for the development of liver fibrosis during chronic liver injury (15,25). Additionally, germ-free mice have reduced hepatocellular tumorigenesis as compared to conventional mice (15). Similar to studies of gastric cancer, antibiotic therapy in mouse models of liver cancer also resulted in significantly reduced liver tumors (15,26,27).
The role of chronic inflammation in tumorigenesis has been well studied in animal models. Evidence in mice has shown that TLR4, a host receptor involved in recognizing microbial patterns, plays a critical role in intestinal microflora mediated hepatocellular carcinoma progression (15,19). Recombinase-activating gene-2-deficient (Rag2–/–) mice which lack functional lymphocytes provide a useful model of chronic inflammatory bowel disease (IBD) and emulate events related to human colon cancer. Infection of Rag2–/– mice with H. hepaticus results in accumulation of neutrophils and macrophages in the colon due to increased tissue inducible nitric oxide (iNOS) (16). This further results in the progression of inflammation and dysplastic changes in the colon leading to cancer. It was shown that by using an inhibitor of iNOS the onset of colon cancer could be completely inhibited in this mouse model (16). Furthermore, the use of T cell transfer paradigm, involving adoptive transfer of proinflammatory CD4+ CD45RB-high T cells to induce IBD in mice not only resulted in increased intestinal polyposis but also surprisingly mammary adenocarcinoma (breast cancer) was observed. Both consequences could be completely abolished by co-transfer of anti-inflammatory CD4+ CD45RB-low regulatory lymphocytes or by neutralization of key proinflammatory cytokine, tumor necrosis factor-alpha (17). These studies indicate that microorganisms contribute to carcinogenesis by promoting inflammation (16,17,24).
Human studies on association of microbiome with cancer
There are at least 10 specific biological agents including bacteria, viruses and parasites that have been implicated in the etiopathogenesis of cancer according to the International Agency for Cancer Research (28). H. pylori infects half of the world population but causes gastric cancer in 1–3% of the population (29,30). Epidemiological analysis of 12 case-control studies has indicated that subjects infected with H. pylori have six times higher risk of non-cardia gastric cancer (31). The relationship between the oral bacterium Fusobacterium nucleatum and colorectal cancer has been observed in human studies. Case-control human cohort studies found higher abundance of Fusobacterium spp. in colorectal adenomas compared with controls (32,33). A reduction in gastric cancer in antibiotic treated patients infected with H. pylori has been demonstrated (34,35) and H. pylori eradication by antibiotics in patients resulted in the regression of gastric mucosa-associated lymphoid tissue (MALT) tumors (36). Uncontrolled adaptive immune responses in patients with chronic infection with H. pylori, Campylobacter jejuni, Borellia burgdorferi, or Chlamydia psittaci may contribute to the development of esophageal cancer, gastric MALT lymphoma, skin MALT lymphoma, and ocular adnexal lymphoma as indicated by epidemiologic studies (34,35,37-41). Toxins produced by enterotoxigenic Bacteroides fragilis have been associated with acute IBD and colorectal neoplasia, especially in late-stage colorectal cancer in humans (42,43). A recent Barrett’s esophagus cohort study found an association between the ratio of Streptococcus to Prevotella taxons and abdominal obesity as well as hiatal hernia length, two known esophageal adenocarcinoma risk factors in Barrett’s esophagus (44). Additional epidemiological evidence suggests chronic infection with Salmonella enterica subsp. enterica serovar Typhi may play a role in the development of gallbladder cancer (45,46). Recent studies have indicated that intestinal bacteria, which are the main source for the portal vein LPS play a key role in hepatocellular carcinoma (22). Overall human studies provide ample evidence in support of microbial etiopathogenesis in various types of cancer (Table 2).

Full table
From a mechanistic point of view, while several human cancers are linked to a single organism, some, like colon cancer are not. An emerging mechanistic hypothesis is that of the “Alpha Bug” (48). This hypothesis suggests that specific microbes, while not singularly responsible for carcinogenesis, may alter the microbial composition of the mileu in which the epithelial cells exist to lead to carcinogenesis. This hypothesis, in effect, integrates the single microbe and microbial communities’ points of view of disease causation.
Assessing the bacterial microbiome
In the majority of investigations summarized above, investigators worked with one microbial species to establish either a strong association or causation. Often, these investigations were facilitated by the ability to culture and experimentally manipulate this species in functionally relevant experiments. However, we now know that the microbiome at every human site is much more complex. Therefore, current assessments of the microbiome rely on more modern methods of identification, especially by 16S ribosomal profiling. In order to interpret these studies, a rudimentary understanding of the technology and methods used in these experiments is helpful.
The approximately 1.5 kb-long 16S ribosomal RNA (rRNA) is a component of the bacterial ribosome, and all bacteria have one or more copies of the 16S gene that generates this rRNA. While the 16S gene is highly conserved across bacterial species, it has nine hypervariable regions (V1–V9) of about 30–100 bp whose sequences are taxon-specific and useful for identifying bacteria to varying taxonomic ranks or levels, from phylum and class onwards possibly up to species This is the basis for microbiome profiling of human samples by 16S amplicon sequencing (Figure 1).
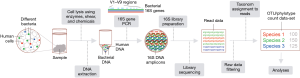
The first step in microbial profiling is isolation of DNA from a sample so that DNA of 16S genes in it can be amplified by polymerase chain reaction (PCR) and subjected to sequencing to identify the microbes that they originate from. Enzymes (49) such as proteinase K and lysozyme, and mechanical means (50) such as beating with sub-millimeter-sized metallic beads or sonication are generally used together to lyse tissue or cells in fluid samples to ensure release of DNA of thick-walled microbes so that it gets collected when DNA is isolated from a lysate. It should be noted that processing of a sample before DNA is extracted from it, such as centrifugation of fluid samples to remove debris, which can cause microbial loss, and formalin-fixation of tissues, which degrades DNA, can have significant impact on its microbial profiling. The method to extract DNA from samples too can affect microbial analysis (51). Exogenous bacteria, or bacterial DNA, can get introduced into samples or their preparations in the 16S sequencing workflow because of contamination from containers, reagents, etc., even if they are bacterially sterile. Inclusion of controls for such contamination points, such as an empty specimen vial or an unused swab, as well as inclusion of a well-defined mock bacterial community sample are important good practices alongside adherence to uniformity across all samples for various steps of the 16S workflow for any microbiome profiling study (52).
The DNA that is isolated from a sample has both host (human) and microbial DNA. The latter is present as a large fraction of the overall DNA content in microbe rich samples such as saliva and bronchioalveolar lavage fluid, or small and minimal for microbe poor samples such as lung parenchyma. This microbial load dictates the number of PCR cycles that is required to amplify the 16S DNA to an amount that is adequate for sequencing. The primers used for PCR target highly conserved regions of 16S such that the PCR amplicon spans one or more of the taxonomic information-bearing V1–V9 16S regions. The V3–V4 16S segment is an amplicon that is used commonly (53). A PCR-amplified 16S product is processed through molecular methods to generate a library that is suitable to undergo single-molecule sequencing. The processing is primarily to enable sequencing (sequencing adapter addition) and to allow for sequencing of multiple libraries in the same instrument run (indexing or barcoding). MiSeq and Ion Torrent instruments manufactured by Illumina® and Thermo Fisher® are widely used for 16S sequencing. From a few to millions of single-molecule 16S DNA reads are generated for a sample in sequencing, depending on the sample’s bacterial load and on the design of the sequencing run, which is usually configured to obtain a few tens of thousands of reads per library. Deeper sequencing (more reads) allows for observing rarer bacterial species but it increases experiment cost.
The principal task in analyzing the sequencing data is to identify the bacterial species/taxons that each of the 16S sequencing reads originates from. Open-source and well-documented software like QIIME and mothur are available for this purpose (54), as well as to perform other tasks such as filtering the sequencing data of poor-quality reads and chimeric 16S reads that can arise during PCR. Sequencing reads are generally binned by this software into operational taxonomic units (OTUs), with groups of reads with similarity greater than a certain threshold (typically 97%) put in one out (55). Reference 16S sequence databases (56) such as Greengenes, SILVA and RDP are then used to assign bacterial information to an OTU to a depth that, depending on the OTU, may be down to the species or even strain taxonomic level or may be shallower and limited to a level such as family or genus. This taxonomic or phylotype assignment maybe the same for multiple OTUs. The final output of the software essentially is count data for each OTU or phylotype that is detected for a sample.
The OTU or phylotype count data-set obtained in a study is further analyzed in different ways depending on the study’s purpose. Common analyses include examining samples for their microbial OTU proportional composition, intra-sample richness for OTUs (alpha diversity), and for inter-sample species differences (beta diversity). A number of metrics, such as Shannon index and chao1 for alpha diversity, and UniFrac distance and Jaccard index for beta diversity, are used when scoring these diversities. Analyses for differential or relative abundance of specific microbial taxa between two sample-groups, and for correlating OTU abundances with a variable of interest requires the use of appropriate data normalization method and statistical test (57). Besides these taxonomical analyses of microbiome 16S sequencing data, functional analyses, such as for metabolic profiles and genetic pathways, and network or ecological relationships at the whole bacterial community or metagenomic level can also be performed using the OTU/phylotype count data-set (58).
Oral microbiome and cancer
As the lung is directly communicating with the oral cavity, the oral microbiome is highly relevant to the lung microbiome. The oral microbiome is a diverse community that includes bacteria, fungi, as well as viruses. The shift from healthy to disease causing microorganisms within the oral microbiome leads to oral diseases like dental caries, periodontal disease and cancer (59). In addition, the oral microbiome has been associated with systemic diseases including infective endocarditis, rheumatoid arthritis, and pulmonary disease (60). More recently there has been a renewed interest in the association of oral microbiome with cancer. There is now evidence of relationship between cancer and tooth loss or periodontal disease (61). Additional studies have shown that there is an association of periodontal disease with hematological, breast and prostate cancers (62). In large epidemiological studies, increase in the relative risk of pancreatic (63) and lung cancer (64) have been documented when associations were studied with periodontal disease. However, because these cancers are multifactorial and mechanistically complex, confounding factors that may influence cancer causation exist. Examples of such factors include but are not limited to smoking, socioeconomic status, demographics, and diet (65). The oral microbiome can aid in conversion of alcohol and smoking byproducts to mutagenic compounds [e.g., alcohol to acetaldehyde and hydroxylation of nitrosamines derived from tobacco smoking (66)]. Epidemiologic studies have demonstrated a relationship between periodontal disease, tooth loss and increased risk of pancreatic cancer. Whether this is a true association or a result of confounders that are common to both complex chronic diseases—periodontitis and cancer, is a challenging question to answer (65). From a microbiome standpoint, the evidence is evolving and studies have shown oral dysbiosis in pancreatic cancer patients. A DNA microarray designed for the oral microbiome compared oral (salivary) microbiome between pancreatic cancer, chronic pancreatitis and controls. It was noted that Neisseria elongata and Streptococcus mitis were significantly decreased in pancreatic cancer patients. Granulicatella adiacens was significantly higher and Streptococcus mitis significantly lower in chronic pancreatitis patients (67). In a prospective evaluation of salivary microbiome in pancreatic adenocarcinoma and matched controls, Porphyromonas gingivalis and Aggregatibacter actinomycetemcommitans were associated with higher risk of cancer development. Conversely, phylum Fusobacteria and genus Leptotrichia were associated with decreased risk of pancreatic cancer (68). With respect to the lung, microaspiration of oral fluids is thought to seed the lungs with oral bacteria (69). In a study using broncheoalveolar lavage samples from 49 healthy subjects it was found that enrichment of the oral taxa in the lungs was associated with a Th17 inflammatory phenotype (69)
Lung microbiome and cancer
Traditionally, the lung was considered a sterile space. Recently, it has been suggested that a lung microbiome exists and alterations in the lung microbiome is associated with disease states such as exacerbations in chronic obstructive pulmonary disease (COPD) (70,71). However, whether a lung cancer tumor microbiome exists was unclear when D’Journo et al. performed PCR for 16S rRNA gene in patients undergoing lung surgery and did not find any bacterial presence, whereas a number of other investigators did (72-74). In their study D’Journo et al. performed a careful bronchoalveolar lavage from the resected specimen from the distal airways in a sterile fashion. PCR was performed for 16S rRNA. Only 6 of 87 patients had a positive amplicon. Sequencing of these amplicons proved they were of human origin. Interestingly, these investigators found that 15% of these patients had Cytomegalovirus (CMV) that could be sequenced from the same specimens and that this was associated with post-operative pneumonia. In contrast, Yu et al. performed 16S sequencing on DNA obtained from the non-cancerous portion of the lung from 165 patients with lung cancer. They not only found a microbiome distinct from that of other body sites, but described associations between specific microbial diversity patterns and epidemiologic exposures. They also found associations between stages of disease with microbial composition, raising interesting mechanistic hypotheses. Similarly, Lee et al. performed bronchoalveolar lavage by bronchoscopy in 28 patients, 20 with lung cancer and 8 without. Not only did they find a large number of OTUs in each sample, they found statistically different OTU abundance in patients with and without lung cancer in even this relatively small sample size. Although obvious, it is important to note that lungs are constantly exposed to microbiota from the inhaled air and the upper respiratory tract. Various lung conditions like COPD, cystic fibrosis, and bronchiectasis are associated with variable microbiota (75). The oral microbiota has direct access to the lung by aspiration. Studies of aspiration pneumonia patients have found an increase in oral streptococci in the bronchoalveolar lavage fluids (76). A recent study has determined that the oral microorganisms Veillonella and Capnocytophaga were found to be significantly higher in the saliva samples of lung cancer patients and that this may be used as a biomarker for early detection of lung cancer (47). A recent study by Greathouse et al. (77) examined the presence of a lung tissue microbiome in 33 patients without lung cancer and 142 patients with lung cancer and found a distinct lung microbiome in patients with lung cancer. These characteristics of the lung cancer microbiome were also seen in the TCGA (The Cancer Genome Atlas) data as well using a custom data analysis pipeline. Thus, there seems to be little doubt that there is a microbiome in both normal lungs as well as in lung cancer. Additional indirect evidence for the relationship between the microbiome and lung cancer is the epidemiologic study by Zhang et al. (78). In this interesting nested case control study, the authors examined the association between significant antibiotic use (>10 prescriptions) and the incidence of lung cancer [risk ratio (RR) 1.3 after adjustment of confounders]. While this may reflect the inflammatory effects of repeat infections it may also be due to changes in the composition of the lung microbiome due to the antibiotics themselves. The latter mechanism is supported by the work of Cheng et al. (79). In this interesting study, the investigators found that administration of antibiotics to mice with Lewis lung cancer tumors shortened their median survival and led to the formation of larger tumors, probably by reducing the γδT17 anti-tumor response.
There are three potential mechanisms for the carcinogenic potential of the lung microbiome:
- Creation of an inflammatory milieu that promotes carcinogenesis. For a large proportion of other cancers, the etiopathogenesis is complex and multifactorial and may include a susceptible host, environmental factors, burden from chronic diseases, habits such as smoking and alcohol consumption and other yet unidentified etiologies (80). Chronic inflammation may account for up to 20% of all cancers and the dysbiotic microbiota may have a role in propagating and sustaining a chronic inflammatory milieu. In cancer of the gastrointestinal system for example, a preceding chronic inflammatory environment has been hypothesized to play a role in carcinogenesis (81). Human cells interact with the outside environment via membrane receptors and transfer message internally for a response through signal transduction. Although this process is complex, largely it is aimed at cell’s functional adaptation to a dynamic external environment. In the event of a microbial insult, the immune system is usually capable of an effective response. However, in situations where the response is chronic, the sustained collateral changes in the microenvironment may be undesirable (80,82-84). Membrane receptors such as pattern-recognition receptor (PRR), cluster of designation (CD), and toll-like receptor (TLR) proteins and others can recognize microbes, microbial products, pro-inflammatory cytokines, signaling molecules and altered human proteins and nucleic acids. Recognition of some of these extracellular molecular signals may result in downstream effects on apoptosis, cell cycle regulation and cellular proliferation. Mutations can result from direct exposure to microbial toxins, microbial oncogenes, reactive oxygen species produced during inflammation and damage to cellular repair mechanisms. Survival of cells that have undergone mutations, their selection and propagation can result in carcinogenesis (80,82-84). The immune effect of the microbiome on lung cancer may be due to specific compositions of both lung and gut microbiomes and deserves further study.
- Metabolic effects of dysbiosis. The microbiome impacts host metabolism and this impact has been amply demonstrated in the gut. Microbial changes have been associated with generation of the carcinogen acetaldehyde (85) as well as deoxycholic acid (86), a bile salt metabolite thought to be involved in esophageal cancer as well as liver cancer. It is entirely possible that such a metabolic imbalance can lead to the formation of toxic metabolites in the lung or may be responsible for secondary processing of procarcinogens in cigarette smoke.
- Genotoxicity. Several bacterial molecules have been associated with genotoxicity. For example, the Bacteroides fragilis toxin has been associated with triggering double stranded DNA damage (87). Chemicals generated by bacteria such as superoxide dismutase is also responsible for genomic instability (88). The study by Greathouse et al. mentioned above also show an association between tumors with TP53 mutations and the presence of Acidovorax. Whether this association is causative or not remains to be seen.
Therefore, there are a number of plausible mechanisms by which the microbiome can lead to or enable carcinogenesis. In addition, the same mechanisms can impact the clinical course of the cancer as well as response to therapy. One association that has garnered significant attention is the relationship between the gut microbiome and response to immunotherapy. A landmark study by Routy et al. (89) demonstrated that patients who respond to immunotherapy with checkpoint inhibitors had a gut microbiome distinct from that of patients who did not. When a fecal transplant was performed from patients who responders to germ free mice, this impacted response to immunotherapy in mice positively compared to fecal transplant from non-responders, favoring a causative role vs. simply an association. The mechanistic underpinnings of this phenomenon were explored by the investigators and is currently the focus of study of several research groups.
Conclusions
The human and oral microbiomes and their various communities of microorganisms play important roles in regulating host functions. Compelling human and animal models provide evidence that microbiota may enhance cancer development in response to the host’s ever-changing internal and environmental factors. The lung has a microbiome and so do lung cancers. While the omics revolution has enhanced our ability to study the lung cancer microbiome, care has to be exercised in the conduct and interpretation of these experiments due to the presence of contaminants as well as the difficulty in proving causation. However, promising early data supports extensive study of the lung cancer microbiome in order to assess the potential for harnessing this knowledge to enhance the therapeutic armamentarium for treating this deadly disease.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lederberg J MA. ’Ome Sweet ’Omics—a genealogical treasury of words. Scientist 2001;15:1.
- Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. [Crossref] [PubMed]
- Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800-12. [Crossref] [PubMed]
- Rous P. A Sarcoma Of The Fowl Transmissible By An Agent Separable From The Tumor Cells. J Exp Med 1911;13:397-411. [Crossref] [PubMed]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer 2010;10:878-89. [Crossref] [PubMed]
- Marshall BJ, Windsor HM. The relation of Helicobacter pylori to gastric adenocarcinoma and lymphoma: pathophysiology, epidemiology, screening, clinical presentation, treatment, and prevention. Med Clin North Am 2005;89:313-44. [Crossref] [PubMed]
- Mager DL. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med 2006;4:14. [Crossref] [PubMed]
- Benamrouz S, Conseil V, Creusy C, et al. Parasites and malignancies, a review, with emphasis on digestive cancer induced by Cryptosporidium parvum (Alveolata: Apicomplexa). Parasite 2012;19:101-15. [Crossref] [PubMed]
- Sacksteder MR. Occurrence of Spontaneous Tumors in the Germfree F344 Rat. J Natl Cancer Inst 1976;57:1371-3. [Crossref] [PubMed]
- Reddy BS, Weisburger J, Narisawa T, et al. Colon carcinogenesis in germ-free rats with 1, 2-dimethylhydrazine and N-methyl-N’-nitro-N-nitrosoguanidine. Cancer Res 1974;34:2368-72. [PubMed]
- Laqueur GL, Matsumoto H, Yamamoto RS. Comparison of the carcinogenicity of methylazoxymethanol-β-D-glucosiduronic acid in conventional and germfree Sprague-Dawley rats. J Natl Cancer Inst 1981;67:1053-5. [PubMed]
- Uronis JM, Mühlbauer M, Herfarth HH, et al. Modulation of the Intestinal Microbiota Alters Colitis-Associated Colorectal Cancer Susceptibility. PLoS ONE 2009;4:e6026. [Crossref] [PubMed]
- Vannucci L, Stepankova R, Kozakova H, et al. Colorectal carcinogenesis in germ-free and conventionally reared rats: Different intestinal environments affect the systemic immunity. Int J Oncol 2008;32:609-17. [PubMed]
- Lee CW, Rickman B, Rogers AB, et al. Helicobacter pylori Eradication Prevents Progression of Gastric Cancer in Hypergastrinemic INS-GAS Mice. Cancer Res 2008;68:3540-8. [Crossref] [PubMed]
- Dapito DH, Mencin A, Gwak GY, et al. Promotion of Hepatocellular Carcinoma by the Intestinal Microbiota and TLR4. Cancer Cell 2012;21:504-16. [Crossref] [PubMed]
- Erdman SE, Rao VP, Poutahidis T, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A 2009;106:1027-32. [Crossref] [PubMed]
- Rao VP, Poutahidis T, Ge Z, et al. Proinflammatory CD4+CD45RBhiLymphocytes Promote Mammary and Intestinal Carcinogenesis inApcMin/+Mice. Cancer Res 2006;66:57-61. [Crossref] [PubMed]
- Reddy BS, Narisawa T, Wright P, et al. Colon carcinogenesis with azoxymethane and dimethylhydrazine in germ-free rats. Cancer Res 1975;35:287-90. [PubMed]
- Fox JG, Wang TC. Helicobacter pylori—not a good bug after all. N Engl J Med 2001;345:829-32. [Crossref] [PubMed]
- Israel DA, Peek RM. Pathogenesis of Helicobacter pylori-induced gastric inflammation. Aliment Pharmacol Ther 2001;15:1271-90. [Crossref] [PubMed]
- Lofgren JL, Whary MT, Ge Z, et al. Lack of Commensal Flora in Helicobacter pylori–Infected INS-GAS Mice Reduces Gastritis and Delays Intraepithelial Neoplasia. Gastroenterology 2011;140:210-20. [Crossref] [PubMed]
- Roderburg C, Luedde T. The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes 2014;5:441-5. [Crossref] [PubMed]
- Garcia-Tsao G, Lee FY, Barden GE, et al. Bacterial translocation to mesenteric lymph nodes is increased in cirrhotic rats with ascites. Gastroenterology 1995;108:1835-41. [Crossref] [PubMed]
- Ward JM, Fox JG, Anver MR, et al. Chronic Active Hepatitis and Associated Liver Tumors in Mice Caused by a Presistent Bacterial Infection With a Novel Helicobacter Species. J Natl Cancer Inst 1994;86:1222-7. [Crossref] [PubMed]
- Seki E, De Minicis S, Österreicher CH, et al. TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat Med 2007;13:1324. [Crossref] [PubMed]
- Schreiber H, Nettesheim P, Lijinsky W, et al. Induction of lung cancer in germfree, specific-pathogen-free, and infected rats by N-nitrosoheptamethyleneimine: enhancement by respiratory infection. J Natl Cancer Inst 1972;49:1107-14. [PubMed]
- Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013;499:97-101. [Crossref] [PubMed]
- de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 2012;13:607-15. [Crossref] [PubMed]
- Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer 2006;118:3030-44. [Crossref] [PubMed]
- Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med 2002;347:1175-86. [Crossref] [PubMed]
- Helicobacter and Cancer Collaborative Group. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut 2001;49:347-53. [Crossref] [PubMed]
- Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 2012;22:292-8. [Crossref] [PubMed]
- McCoy AN, Araújo-Pérez F, Azcárate-Peril A, et al. Fusobacterium is associated with colorectal adenomas. PloS One 2013;8:e53653. [Crossref] [PubMed]
- Ma JL, Zhang L, Brown LM, et al. Fifteen-Year Effects of Helicobacter pylori, Garlic, and Vitamin Treatments on Gastric Cancer Incidence and Mortality. J Natl Cancer Inst 2012;104:488-92. [Crossref] [PubMed]
- Wong BC, Lam SK, Wong WM, et al. Helicobacter pylori Eradication to Prevent Gastric Cancer in a High-Risk Region of China. JAMA 2004;291:187. [Crossref] [PubMed]
- Wotherspoon AC, Diss TC, Pan L, et al. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 1993;342:575-7. [Crossref] [PubMed]
- Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest 2007;117:60-9. [Crossref] [PubMed]
- Islami F, Kamangar F. Helicobacter pylori and Esophageal Cancer Risk: A Meta-analysis. Cancer Prev Res (Phila) 2008;1:329-38. [Crossref] [PubMed]
- Lecuit M, Abachin E, Martin A, et al. Immunoproliferative Small Intestinal Disease Associated withCampylobacter jejuni. N Engl J Med 2004;350:239-48. [Crossref] [PubMed]
- Peek RM, Blaser MJ. Helicobacter Pylori And Gastrointestinal Tract Adenocarcinomas. Nat Rev Cancer 2002;2:28-37. [Crossref] [PubMed]
- Senff NJ, Noordijk EM, Kim YH, et al. European Organization for Research and Treatment of Cancer and International Society for Cutaneous Lymphoma consensus recommendations for the management of cutaneous B-cell lymphomas. Blood 2008;112:1600-9. [Crossref] [PubMed]
- Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015;60:208-15. [Crossref] [PubMed]
- Prindiville TP, Sheikh RA, Cohen SH, et al. Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg Infect Dis 2000;6:171. [Crossref] [PubMed]
- Gall A, Fero J, McCoy C, et al. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLoS One 2015;10:e0129055. [Crossref] [PubMed]
- Caygill CPJ, Hill MJ, Braddick M, et al. Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet 1994;343:83-4. [Crossref] [PubMed]
- Welton JC, Marr JS, Friedman SM. Association Between Hepatobiliary Cancer And Typhoid Carrier State. Lancet 1979;1:791-4. [Crossref] [PubMed]
- Yan X, Yang M, Liu J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res 2015;5:3111. [PubMed]
- Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis 2011;203:306-11. [Crossref] [PubMed]
- Gill C, van de Wijgert JH, Blow F, et al. Evaluation of Lysis Methods for the Extraction of Bacterial DNA for Analysis of the Vaginal Microbiota. PLoS One 2016;11:e0163148. [Crossref] [PubMed]
- Vandeventer PE, Weigel KM, Salazar J, et al. Mechanical disruption of lysis-resistant bacterial cells by use of a miniature, low-power, disposable device. J Clin Microbiol 2011;49:2533-9. [Crossref] [PubMed]
- Kennedy NA, Walker AW, Berry SH, et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One 2014;9:e88982. [Crossref] [PubMed]
- Kim D, Hofstaedter CE, Zhao C, et al. Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017;5:52. [Crossref] [PubMed]
- Takahashi S, Tomita J, Nishioka K, et al. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One 2014;9:e105592. [Crossref] [PubMed]
- Mysara M, Njima M, Leys N, et al. From reads to operational taxonomic units: an ensemble processing pipeline for MiSeq amplicon sequencing data. Gigascience 2017;6:1-10. [Crossref] [PubMed]
- Nguyen NP, Warnow T, Pop M, et al. A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes 2016;2:16004. [Crossref] [PubMed]
- Balvočiūtė M, Huson DH. SILVA, RDP, Greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genomics 2017;18:114. [Crossref] [PubMed]
- Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017;5:27. [Crossref] [PubMed]
- Mukherjee A, Chettri B, Langpoklakpam JS, et al. Bioinformatic Approaches Including Predictive Metagenomic Profiling Reveal Characteristics of Bacterial Response to Petroleum Hydrocarbon Contamination in Diverse Environments. Sci Rep 2017;7:1108. [Crossref] [PubMed]
- Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett 2014;162:22-38. [Crossref] [PubMed]
- Maddi A, Scannapieco FA. Oral biofilms, oral and periodontal infections, and systemic disease. Am J Dent 2013;26:249-54. [PubMed]
- Nwizu NN, Marshall JR, Moysich K, et al. Periodontal Disease and Incident Cancer Risk among Postmenopausal Women: Results from the Women’s Health Initiative Observational Cohort. Cancer Epidemiol Biomarkers Prev 2017;26:1255-65. [Crossref] [PubMed]
- Dizdar O, Hayran M, Guven DC, et al. Increased cancer risk in patients with periodontitis. Curr Med Res Opin 2017;33:2195-200. [Crossref] [PubMed]
- Michaud DS, Joshipura K, Giovannucci E, et al. A Prospective Study of Periodontal Disease and Pancreatic Cancer in US Male Health Professionals. J Natl Cancer Inst 2007;99:171-5. [Crossref] [PubMed]
- Hujoel PP, Drangsholt M, Spiekerman C, et al. An Exploration of the Periodontitis–Cancer Association. Ann Epidemiol 2003;13:312-6. [Crossref] [PubMed]
- Meyer MS, Joshipura K, Giovannucci E, et al. A review of the relationship between tooth loss, periodontal disease, and cancer. Cancer Causes Control 2008;19:895-907. [Crossref] [PubMed]
- Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 2012;23:399-404. [Crossref] [PubMed]
- Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012;61:582-8. [Crossref] [PubMed]
- Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67:120-7. [Crossref] [PubMed]
- Segal LN, Clemente JC, Tsay JC, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol 2016;1:16031. [Crossref] [PubMed]
- Huang YJ, Sethi S, Murphy T, et al. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol 2014;52:2813-23. [Crossref] [PubMed]
- Mammen MJ, Sethi S. COPD and the microbiome. Respirology 2016;21:590-9. [Crossref] [PubMed]
- D’Journo XB, Bittar F, Trousse D, et al. Molecular detection of microorganisms in distal airways of patients undergoing lung cancer surgery. Ann Thorac Surg 2012;93:413-22. [Crossref] [PubMed]
- Lee SH, Sung JY, Yong D, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 2016;102:89-95. [Crossref] [PubMed]
- Yu G, Gail MH, Consonni D, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol 2016;17:163. [Crossref] [PubMed]
- Dickson RP. The microbiome and critical illness. Lancet Respir Med 2016;4:59-72. [Crossref] [PubMed]
- Akata K, Yatera K, Yamasaki K, et al. The significance of oral streptococci in patients with pneumonia with risk factors for aspiration: the bacterial floral analysis of 16S ribosomal RNA gene using bronchoalveolar lavage fluid. BMC Pulm Med 2016;16:79. [Crossref] [PubMed]
- Greathouse KL, White JR, Vargas AJ, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol 2018;19:123. [Crossref] [PubMed]
- Zhang H, Garcia Rodriguez LA, Hernandez-Diaz S. Antibiotic use and the risk of lung cancer. Cancer Epidemiol Biomarkers Prev 2008;17:1308-15. [Crossref] [PubMed]
- Cheng M, Qian L, Shen G, et al. Microbiota modulate tumoral immune surveillance in lung through a gammadeltaT17 immune cell-dependent mechanism. Cancer Res 2014;74:4030-41. [Crossref] [PubMed]
- Elinav E, Nowarski R, Thaiss CA, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 2013;13:759-71. [Crossref] [PubMed]
- Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med 2000;248:171-83. [Crossref] [PubMed]
- Grivennikov SI, Greten FR, Karin M. Immunity, Inflammation, and Cancer. Cell 2010;140:883-99. [Crossref] [PubMed]
- Karin M, Lawrence T, Nizet V. Innate Immunity Gone Awry: Linking Microbial Infections to Chronic Inflammation and Cancer. Cell 2006;124:823-35. [Crossref] [PubMed]
- Kipanyula MJ, Seke Etet PF, Vecchio L, et al. Signaling pathways bridging microbial-triggered inflammation and cancer. Cell Signal 2013;25:403-16. [Crossref] [PubMed]
- Vanhaecke L, Knize MG, Noppe H, et al. Intestinal bacteria metabolize the dietary carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine following consumption of a single cooked chicken meal in humans. Food Chem Toxicol 2008;46:140-8. [Crossref] [PubMed]
- Keren N, Konikoff FM, Paitan Y, et al. Interactions between the intestinal microbiota and bile acids in gallstones patients. Environ Microbiol Rep 2015;7:874-80. [Crossref] [PubMed]
- Cuevas-Ramos G, Petit CR, Marcq I, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 2010;107:11537-42. [Crossref] [PubMed]
- Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, et al. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 2012;3:448. [Crossref] [PubMed]
- Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91-7. [Crossref] [PubMed]


