Circulating free tumor-derived DNA to detect EGFR mutations in patients with advanced NSCLC: French subset analysis of the ASSESS study
Introduction
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) are standard-of-care first-line therapy for patients with advanced non-small cell lung cancer (NSCLC), with tumors harboring activating EGFR mutations. The success of TKIs in these patients is supported by findings from several clinical trials that have demonstrated superior efficacy when comparing TKIs with platinum-based doublet-chemotherapy in large phase III studies (1-6). Based on their demonstrated efficacy, clinical guidelines currently recommend that all patients with advanced NSCLC are tested for EGFR mutations and EGFR mutation-positive patients prioritized for TKI therapy (7). While tissue/cytology samples are the preferred choice for EGFR mutation testing, around 85% of patients with NSCLC are diagnosed at the advanced stages of disease. As a result, tissue/cytology samples are normally obtained via minimally invasive techniques, restricting the quality and quantity of sampling material (8). To overcome this limitation, research has since focused on identifying alternative sample sources for patients with advanced NSCLC, including from plasma (1,8,9), urine (10,11), and spinal fluid (12-14).
In clinical trials of patients with NSCLC, plasma-derived circulating free tumor-derived DNA (ctDNA) has been shown to be a feasible sample source for EGFR mutation testing (1). Moreover, in patients with NSCLC, the presence of EGFR mutations in ctDNA has also shown to predict therapeutic response to TKI treatment (5,8). Although the use of ctDNA analysis in real-world clinical practice is appealing, limited data currently exist for its accuracy, suitability, and feasibility outside the clinical trial setting.
Recently, ASSESS, a multicenter, non-interventional, diagnostic study (NCT01785888), investigated the concordance of EGFR mutation status between matched tissue/cytology and plasma ctDNA samples in patients with advanced NSCLC, in a real-world clinical setting (15). Although this study identified that the concordance of mutation status in 1,162 matched samples was 89% [sensitivity 46%, specificity 97%, positive predictive value (PPV) 78% and negative predictive value (NPV) 90%], different standardized test methodologies were applied across different countries, resulting in variability in EGFR mutation sensitivity values (36–100%) (15). To account for this variability, it is key that subset analyses in different countries are performed to allow for a standardized assessment of the concordance of EGFR mutation status. Here we present data from a subset analysis of all patients who were enrolled in the ASSESS study across 9 centers in France.
Methods
Study design and participants
Full details of the ASSESS study have previously been published (15). For the French subset analysis, all patients were ≥18 years of age with either histologically/cytologically confirmed, treatment-naive, locally advanced NSCLC (stage IIIA/B according to the 7th American Joint Committee on Cancer staging system) that was unsuitable for curative treatment or chemoradiotherapy, metastatic disease (stage IV), or recurrent disease after previous curative treatment (surgical resection and/or adjuvant chemotherapy). All patients provided diagnostic tissue/cytology samples and a 10-mL blood sample for plasma extraction at study enrollment and provided written, informed consent. Study approval was obtained from the independent ethics committees at each institution [French sites: Comité de protection des personnes (Reference No 2013-A01659-34) and Agence nationale du sécurité du medicament et des produits de santé (Reference No 130197B-12)], and the study was conducted in accordance with the Declaration of Helsinki, the International Conference on Harmonization/Good Clinical Practice, applicable regulatory requirements for non-interventional studies, and AstraZeneca’s policy on bioethics and human biological samples.
Outcomes
The primary outcome of this subset analysis was the concordance of EGFR mutation status between matched tissue/cytology and plasma ctDNA samples, including the sensitivity, specificity, PPV, and NPV of EGFR mutation status. Other outcomes were EGFR mutation testing practices, including testing methodologies, sample types, and mutation frequency, and the correlation between mutation and demographic data/disease status.
Procedures
Prior to enrollment, EGFR mutation test data and results for tumor samples were collected where available. Throughout the study, diagnostic tissue/cytology samples underwent EGFR mutation testing as per local practices, after histopathological review [World Health Organization (WHO) classification (16)]. For ctDNA plasma testing, all patients provided a 10-mL blood sample which was collected in EDTA tubes. Plasma was collected from blood samples within 4 h, frozen at −80 °C, transported, and analyzed for EGFR mutations within a single designated laboratory in France. DNA extraction and EGFR mutation testing was performed by allele refractory mutant specific amplification (ARMS) using the Qiagen Therascreen®EGFR Rotor-Gene Q (RGQ) polymerase chain reaction (PCR) kit, as previously described (9,17,18).
Statistical analyses
The concordance of EGFR mutation status between matched tissue/cytology and plasma samples was calculated for the evaluable population [all eligible patients with known tumor (tissue/cytology) and plasma sample mutation status] via concordance rate, sensitivity, specificity, PPV, and NPV, and exact two-sided 95% confidence intervals (CIs). The sample size of the ASSESS study was calculated as follows: for 100 patients with EGFR mutation-positive NSCLC, if the sensitivity was 50%, the 95% CI would be expected to be 40–60. Assuming a 10% prevalence of EGFR mutations in Europe, it was estimated that 1,000 patients should be tested to obtain 100 patients with EGFR mutation-positive NSCLC (16). EGFR mutation testing practices (enrolled population) and EGFR mutation frequency (evaluable tumor/plasma populations) for the French subset of patients were summarized using appropriate descriptive statistics. The correlation between EGFR mutation status and demographic characteristics or disease status was analyzed using a multivariate logistic regression model using stepwise forward selection (10% significance level for entry into the model), constructed to model EGFR tumor mutation status at baseline (EGFR mutation positive versus EGFR mutation negative from tumor sample). Key demographic characteristics included histology (adenocarcinoma versus non-adenocarcinoma), smoking status (never- versus ever-smoked), gender, age (≤65 vs. >65 years of age), and WHO performance score (0–1, 2), and key disease status characteristics including time since diagnosis, current disease status (IIIA versus IIIB versus IV), and number of organs with metastasis (fitted as a continuous covariate). An odds ratio (OR) <1 implied that the patient was more likely to have a negative mutation status if that particular variable was present.
Results
Patient demographics and clinical characteristics
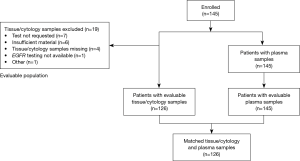
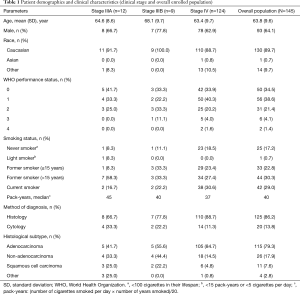
Of 1,311 patients enrolled in the ASSESS study, 145 (11.1%) patients were recruited from 9 centers across France and constituted the enrolled population for all outcome analyses (Figure 1). Last patient study visit was 17th April 2014. Patient demographics and clinical characteristics for the enrolled French population are summarized in Table 1. The mean (standard deviation) patient age was 63.8 (9.6) years and 64.1% of patients were male. The majority of patients (89.7%) were Caucasian, had a WHO performance status of 0 (34.5%) or 1 (38.6%), were former (53.1%) or current (29.0%) smokers, and were diagnosed with adenocarcinoma (79.3%). The most common method of diagnosis was by histopathological examination (86.2%).
Full table
Sampling and EGFR mutation testing methodologies
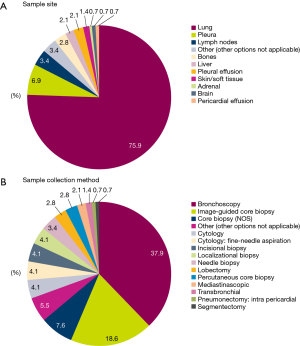
In the majority of patients, tissue/cytology samples were collected from primary tumors in the lung (75.9%) (Figure 2A) using bronchoscopy (37.9%) or image-guided core biopsy (18.6%) (Figure 2B).
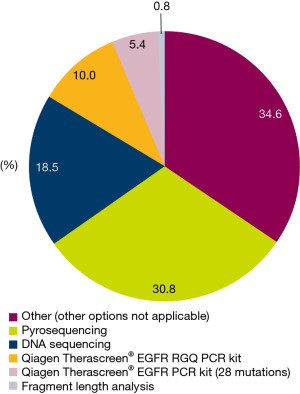
Of the 145 enrolled patients, 126 (86.9%) had evaluable tumor samples. Reasons for non-evaluable samples included insufficient material provided for testing (6/19; 31.6%), EGFR mutation test not requested (7/19; 36.8%), or EGFR mutation testing not available (1/19; 5.3%). In 5 cases, no reason was provided or data were missing (5/19; 26.3%) (Figure 1). The most common EGFR mutation testing methods included pyrosequencing (30.8%), DNA sequencing (18.5%), and ARMS using the Qiagen Therascreen®EGFR RGQ PCR kit (10.0%) (Figure 3). The median turnaround time for the testing of tissue/cytology samples was 13.0 days (range, 2.0–290.0 days) and testing success rate was 96.9%.
For plasma ctDNA testing, all 145 enrolled patients had evaluable samples and all plasma samples were analyzed using the ARMS Qiagen Therascreen®EGFR RGQ PCR kit. Testing success rate was 100%.
EGFR mutation frequency (tissue/cytology and plasma evaluable populations)
The EGFR mutation frequency in evaluable tissue/cytology and plasma samples was 10.3% (13/126 samples) and 6.9% (10/145 samples), respectively (Table 2). The most common EGFR mutations in tissue/cytology and plasma samples included the exon 19 deletion and L858R mutation. EGFR mutation frequencies predominated in patients with tumor, node, metastasis stage IV M1b adenocarcinoma (Table 2).
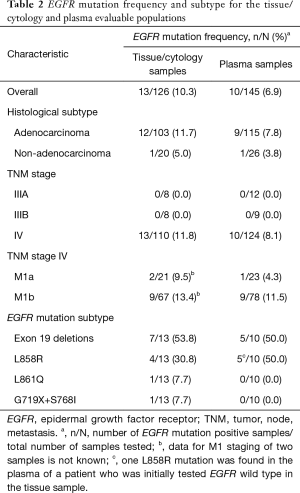
Full table
Correlations between EGFR mutation status and demographic and disease characteristics
EGFR mutation rates were significantly higher in ever- versus never-smokers using stepwise logistic regression for both tumor and plasma testing [tumor: OR =112.7 (95% CI, 12.9–989.4), P<0.0001; plasma: OR =44.0 (95% CI, 4.8–405.2), P=0.0008]. No significant correlation was identified in plasma testing when comparing EGFR mutation rates in male versus females.
EGFR mutation status concordance between matched tissue/cytology and plasma samples (tissue/cytology and plasma evaluable populations)
The concordance of mutation status was measured between 126 matched tissue/cytology and plasma samples. Overall concordance was 96.0% [PPV 100.0% (95% CI, 66.4–100.0%) and NPV 95.7% (95% CI, 90.3–98.6%)] (Table 3). Of the 113 patients who were EGFR mutation-negative in tissue, one sample tested positive in plasma. Reanalysis of the frozen tissue and formalin-fixed paraffin-embedded samples using two different techniques (ARMS and digital PCR) confirmed the presence of a L858R mutation, indicating a tissue false-negative result (mutation not detected by pyrosequencing). Sensitivity was therefore adjusted to 64.3% (95% CI, 35.1–87.2%) and specificity to 100.0% (95% CI, 96.8–100.0%) (Table 3).

Full table
Discussion
To our knowledge, ASSESS is one of the largest studies of real-world EGFR mutation testing practices in Europe and Japan (15). This subset analysis of French patients demonstrated comparable concordance (96.0%) to those previously reported for the overall European and Japanese cohorts in the ASSESS study (92%). Moreover, sensitivity, specificity, PPV, and NPV values were higher in the French subset analysis than those reported for the entire and European cohorts in ASSESS (French ASSESS cohort: sensitivity 64.3%, specificity 100.0%, PPV 100.0%, NPV 95.7%; Overall ASSESS cohort: sensitivity 46%, specificity 97%, PPV 78%, NPV 90%). It should be noted that sensitivity was calculated based on the analysis of a small number of patients (n=14), resulting in a large 95% CI (35.1–87.2). In the French patients, EGFR mutations were only identified in patients with stage IV lung cancer and higher sensitivities for plasma testing were observed in M1b patients (100%, n=9/9) compared with the overall ASSESS cohort (19). This is most likely related to the higher tumor burden of these patients.
With the caveat of cross-trial comparisons, these results are comparable to the open-label IFUM study of Caucasian patients with EGFR mutation-positive NSCLC, in which a mutation status concordance of 94.3% (sensitivity 65.7%, specificity 99.8%, PPV 98.6%, NPV 93.8%) was observed between 652 matched tissue/cytology and plasma samples (20). It must also be highlighted that this study used a similar testing technique to the ASSESS study: the Qiagen QIAamp Circulating Nucleic Acid Kit (20). Several reports suggest that the Qiagen ARMS-based Therascreen®EGFR RGQ PCR kit may not be the most sensitive method available (21,22); however, it has been shown to be robust and was widely used by diagnostic laboratories in 2013, including in the enrolment for the ASSESS study (15). The Qiagen ARMS-based Therascreen®EGFR RGQ PCR kit was also approved by the US Food and Drug Administration in 2013 as companion diagnostic to select patients suitable for EGFR TKI therapy (23).
Within the French subset, no false-positives or -negatives were reported from EGFR mutation plasma testing and only one false-negative was reported from tissue that was tested. It is possible that other false-negatives may have been present but, unfortunately, no further plasma samples were available for subsequent testing with more sensitive techniques, including digital PCR. However, it is worth mentioning that the samples may have given a negative result due to the absence of tumor DNA within the sample. Although plasma testing demonstrated 100% specificity, testing was conducted in a central testing laboratory, while tissue/cytology testing was performed according to local practices. Differences in mutation testing methodologies between different laboratories may therefore account for sensitivity differences between the different sampling types.
Across the enrollment population in the French subset study, approximately 10% of patients did not qualify for tissue/cytology EGFR mutation testing. In these patients, plasma EGFR mutation testing was a feasible option to confirm mutation status and inform suitability for EGFR TKI treatment. However, it should be noted that although this study was pre-planned, the retrospective analysis was performed in a relatively small number of patients. Furthermore, analysis of FFPE samples was performed between April 2013 and April 2014; however, at the current time in France, most of the tissue samples are tested using next-generation sequencing. In the future, it may be of interest to perform large-scale concordance studies for EGFR mutation testing from tumor and plasma samples using standardized sensitive mutation testing methods to ensure accuracy of results.
Taken together, these real-world data imply that ctDNA plasma testing may be used as an alternative, minimally invasive sampling method to assess EGFR mutation status and suitability for EGFR TKI treatment in patients with advanced NSCLC, particularly when tissue biopsy may not be recommended or available. Given that previous studies have demonstrated the predictive value of dynamic ctDNA EGFR mutational changes and response to EGFR TKI treatment (17,18,24,25), advantage could further be taken of this technology to temporally monitor ctDNA as a means of assessing treatment efficacy or success in patients with advanced NSCLC.
Acknowledgements
This work was supported by AstraZeneca and coordinated by Worldwide Clinical Trials, which also managed the database and performed the primary analyses. In collaboration with AstraZeneca, the study steering committee interpreted the study results. Dr Denis had full access to the study data and final responsibility for the decision to submit for publication. This study was sponsored by AstraZeneca. Medical writing support, under the direction of the authors, was provided by Mark Holland PhD of CMC CONNECT, a division of Complete Medical Communications Ltd, Macclesfield, UK, funded by AstraZeneca, Cambridge, UK, in accordance with Good Publication Practice (GPP3) guidelines. We are most grateful to JJ Paillusson for expert technical assistance.
Footnote
Conflicts of Interest: MG Denis: grants/research support/consultant for AstraZeneca, BMS, Boehringer Ingelheim, Merck, Qiagen, Roche Pharma, and Takeda. G Le Garff: clinical trials for AstraZeneca, Lilly, and Roche; all financial contributions except MUTACT study (AstraZeneca) paid by clinical research unit; board member for Novartis; invitation to congress from Air Santé, Altana, AstraZeneca (before 2012), Boehringer Ingelheim, GSK, LFB, Lilly, MSD, Pierre Fabre, and Roche. C Locher: membership on advisory board for AstraZeneca, BMS, Boehringer Ingelheim, Pfizer, and Roche; medical conferences for Novartis. M Licour: employee of AstraZeneca. N Normanno: grants/research support/consultant for AstraZeneca, Qiagen, and Roche Diagnostics. M Reck: member of the speaker’s bureau for AstraZeneca, BMS, Boehringer Ingelheim, Daiichi-Sankyo, Hoffmann-La Roche, Lilly, and Pfizer; consultant for AstraZeneca, BMS, Boehringer Ingelheim, Daiichi Sankyo, Hoffmann-La Roche, Lilly, MSD, and Pfizer. All other authors have declared no conflicts of interest.
Ethical Statement: For the French sites discussed in this manuscript, ethical approval was provided by the Comité de protection des personnes (Reference No: 2013-A01659-34) and the Agence nationale du sécurité du medicament et des produits de santé (Reference No: 130197B-12).
References
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Riely GJ, Politi KA, Miller VA, et al. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res 2006;12:7232-41. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Zhou C, Wu Y-L, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- Fenizia F, De Luca A, Pasquale R, et al. EGFR mutations in lung cancer: from tissue testing to liquid biopsy. Future Oncol 2015;11:1611-23. [Crossref] [PubMed]
- Vallée A, Marcq M, Bizieux A, et al. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 2013;82:373-4. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Tchekmedyian N, Mudad R, Blanco FF, et al. Longitudinal monitoring of ctDNA EGFR mutation burden from urine correlates with patient response to EGFR TKIs: A case series. Lung Cancer 2017;108:22-8. [Crossref] [PubMed]
- Théoleyre S, Masson I, Herbreteau G, et al. Treatment of a NSCLC patient with osimertinib based on the detection of the EGFR T790M resistance mutation in cerebrospinal fluid. Lung Cancer 2017;114:111-2. [Crossref] [PubMed]
- Yang H, Cai L, Zhang Y, et al. Sensitive detection of EGFR mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn 2014;16:558-63. [Crossref] [PubMed]
- Zhao J, Ye X, Xu Y, et al. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol 2016;78:1305-10. [Crossref] [PubMed]
- Reck M, Hagiwara K, Han B, et al. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: the ASSESS study. J Thorac Oncol 2016;11:1682-9. [Crossref] [PubMed]
- Brambilla E, Travis WD, Colby TV, et al. The new World Health Organization classification of lung tumours. Eur Respir J 2001;18:1059-68. [Crossref] [PubMed]
- Vallée A, Audigier-Valette C, Herbreteau G, et al. Rapid clearance of circulating tumor DNA during treatment with AZD9291 of a lung cancer patient presenting the resistance EGFR T790M mutation. Lung Cancer 2016;91:73-4. [Crossref] [PubMed]
- Marcq M, Vallee A, Bizieux A, et al. Detection of EGFR mutations in the plasma of patients with lung adenocarcinoma for real-time monitoring of therapeutic response to tyrosine kinase inhibitors? J Thorac Oncol 2014;9:e49-e50. [Crossref] [PubMed]
- Normanno N, Brown H, Haddad V, et al. Clinical and demographic features that influence EGFR mutation detection in plasma from patients (pts) with aNSCLC: the ASSESS experience. J Thorac Oncol 2016;11:S151. [Crossref]
- Douillard J-Y, Ostoros G, Cobo M, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open label, single arm study. Br J Cancer 2014;110:55-62. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Normanno N, Denis MG, Thress KS, et al. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017;8:12501-16. [Crossref] [PubMed]
- Hsiue EH, Lee JH, Lin CC, et al. Profile of the therascreen® EGFR RGQ PCR kit as a companion diagnostic for gefitinib in non-small cell lung cancer. Expert Rev Mol Diagn 2016;16:1251-7. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Zhou Q, Yang JJ, Chen ZH, et al. Serial cfDNA assessment of response and resistance to EGFR-TKI for patients with EGFR-L858R mutant lung cancer from a prospective clinical trial. J Hematol Oncol 2016;9:86. [Crossref] [PubMed]