TMEM213 as a novel prognostic and predictive biomarker for patients with lung adenocarcinoma after curative resection: a study based on bioinformatics analysis
Introduction
Lung cancer is the major cause of cancer death worldwide (1). The vast majority of lung cancer is non-small cell lung cancer (NSCLC), of which adenocarcinoma accounts for 50%, being the most common histological type (2). Currently, the American Joint Committee on Cancer (AJCC) staging system is the primary reference for guiding clinical decisions and is by far the very best predicting factor of prognosis in patients with NSCLC. Over 20% of early-stage patients eventually progress to disease recurrence and metastasis, which suggests that current survival predictions are deficient and in need of improvement (3). This also suggests that occult metastases are present as early as at the time of surgical intervention. Therefore, adjuvant chemotherapy (ACT) is recommended to improve the prognosis of patients who undergo a complete resection. Although the use of adjuvant paclitaxel-carboplatin has not been demonstrated in prospective studies, research has shown that it can be considered as the ACT for resected NSCLC (4-6). Nevertheless, some findings regarding the use of adjuvant paclitaxel-carboplatin have raised controversy. One phase III trial denies the benefit of adjuvant paclitaxel and carboplatin in resected early-stage lung cancer (7). Another study reported that stage II–IIIA patients usually receive platinum-based ACT after surgical resection, but only 4–15% survival benefit has been observed (8). This suggests that NSCLC is very heterogeneous, and this potential heterogeneity is not well-reflected in the current staging system, which significantly confuses treatment of patients. Therefore, it is essential to find a novel and effective prognostic factor to differentiate patients who might benefit from adjuvant paclitaxel-carboplatin and those who are not sensitive to it and need other, more active treatments.
The involvement of transmembrane proteins (TMEMs) in malignancy has recently attracted the interest of researchers. TMEMs are a group of 310 different proteins predicted to be components of cellular membranes, such as lysosomes, mitochondrial membranes, and the Golgi apparatus. The role of most TMEM proteins remains unclear, mainly due to the difficulty in purification and extraction of TMEM proteins (9). Moreover, many TMEM proteins have been functionally designated as transmembranous anion channels (10). Members of TMEMs are frequently abnormally expressed in various cancers, such as hepatocellular carcinoma (TMEM7) (11), gastric carcinoma (TMEM16A) (12), renal cell carcinoma (TMEM22) (13), lymphomas (TMEM176) (14), glioma (TMEM97) (15), and ovarian cancer (TMEM45A and TMEM158) (16,17). TMEM213 belongs to a member of the TMEM family. Kamiński found that TMEM213 SNPs may be involved in the pathogenesis of coronary artery disease (18). Sang Yun analyzed the differences in gene expression between peripheral T-cell lymphoma and normal reactive lymph nodes and found that TMEM213 was significantly up-regulated (19). It has also been reported that significant deregulation of expression of TMEM213 in clear cell renal cell carcinoma (ccRCC) may be involved in tumorigenesis and associated with the invasion and metastasis of ccRCC (9). However, there has been no investigation yet conducted concerning the relationship between TMEM213 expression and clinicopathological features and the prognosis of lung adenocarcinoma.
It has been demonstrated that gene-expression profiles obtained by cDNA microarray analysis can provide detailed features of individualized cancers and such information might improve the clinical tactics for neoplastic diseases by developing novel drugs and providing the foundation of personalized treatment (20). In this study, a screening approach was performed on RNA-Seq expression data from The Cancer Genome Atlas (TCGA) database and microarray datasets from the Gene Expression Omnibus (GEO) database to find a possible target in the TMEM family whose expression could serve as an independent prognostic predictor of lung adenocarcinoma after curative resection and benefit from adjuvant paclitaxel-carboplatin.
Methods
Patient screening process
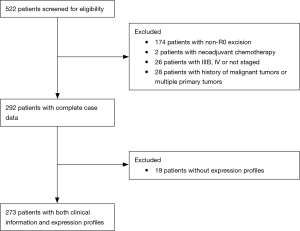
Clinical information of patients with lung adenocarcinoma was obtained from the TCGA database. There were 522 patients screened for eligibility, excluding 174 patients with non-RO excision; 2 patients with neoadjuvant chemotherapy; 26 patients with IIIB, IV or not staged; 28 patients with a history of malignant tumors or multiple primary tumors; and 19 patients without expression profiles. The final 273 patients with both clinical information and expression profiles were included in the analysis (Figure 1).
Screening prognostic genes
Patients with lung adenocarcinoma in the TCGA database were selected, and survival analysis was performed for all genes using the R software (R 3.0.2). Overall survival (OS) was defined by the time from the beginning of surgery to death or the last follow-up date. Using the MAX STAT function package, the log-rank statistic was calculated based on the Conditional-Monte Carlo method. The optimal cut-off value for the predicted prognosis between the 10th and 90th is the binary value for each gene (P<0.05). Based on Univariate COX analysis, genes with P values of <0.05 were selected for the follow-up analysis.
Screening drug-related genes by the Subpopulation Treatment Effect Pattern Plot (STEPP)
In the NCBI GEO datasets, GSE42127 is the dataset related to ACT and prognosis, which was attached as the supplementary information in relevant research (PMID: 23357979) (21). Interactions between ACT and each screened gene were analyzed with STEPP methodology in GSE42127. STEPP is a graphical tool that helps researchers explore the heterogeneity of treatment effects based on the value of a consecutive baseline covariate in overlapping subpopulations (22). Briefly, STEPP uses a sliding-windows method to define several overlapping subpopulations of the patient based on a continuous covariate, such as gene expression, and plots the resulting treatment effects estimated within each subpopulation (23). STEPP analyses were performed using the R (http://cran.r-project.org/) software with Package “STEPP.” Finally, the drug-related gene was screened according to the STEPP pattern and P value.
Statistical analysis
R statistical software (version 3.0.2) was used for statistical analyses. All statistical tests were two-tailed and P values less than 0.05 were considered statistically significant. TMEM213 expression was a binary variable (low/high) for analysis. Other categorical variables included gender, T-stage, N-stage, and TNM stage. Age was a numeric variable. The relationships between TMEM213 expression levels with categorical variables were examined using the Chi-squared test or Fisher’s exact test. Welch’s two independent sample t-test or nonparametric Mann-Whitney U test was used to compute P values in comparing continuous variables. The Kaplan-Meier (KM) survival curve was reported, and the time-to-event data were compared using the log-rank test. Univariate analysis analyzing OS and independent variables, including TMEM213 and other clinic-pathological parameters (CPPs) were conducted using the Cox proportional hazard (PH) regression model. Corresponding hazards ratios (HRs) and 95% confidence interval (CI) were reported. Multivariable Cox regression models were further conducted by adjusting for other CPPs (age, gender, T-stage, and N-stage). TMEM213 was modeled as dichotomous according to the relative fit of multivariate models adjusted for the standard prognostic factors. The stepwise selection was conducted, and the final model was selected based on the Akaike information criterion (AIC). Internal validation was conducted using the bootstrapping method (1,000 replications). Corresponding estimate and 95% CI were also reported.
The external validation was conducted using the online Kaplan-Meier plotter database analysis (24). OS was estimated by median TMEM213 expression for lung adenocarcinoma patients. All other parameters were set at default settings, except for the “treatment group.” It was set for “only surgical margins negative” to replicate our current study cohort conditions best.
Enriched pathway analysis
Gene set enrichment analysis (GSEA) assay was performed to investigate the biological characteristics shared by different TMEM213 expression levels. GSEA was performed using the software GSEA v2.2.2 (www.broadinstitute.org/software/gsea). The TMEM213 expression level was annotated as high or low phenotype, and C2: curated gene setsCP: KEGG: KEGG gene sets from the Molecular Signatures Database (MSigDB) were utilized. All other parameters were set to default.
Results
Patient characteristics
Clinical information of 273 lung adenocarcinoma patients was obtained from publicly available TCGA database (Table 1). The median age was 66 (range, 33–84) years. Males accounted for 46.9% (n=128), females accounted for 53.1% (n=145), and 72.5% were white (n=198). T1/T2 and T3/T4 accounted for 91.2% (n=249) and 8.8% (n=24), respectively. N0/Nx and N1/N2 accounted for 68.9% (n=188) and 31.1% (n=85), respectively. Stage I and II/IIIA accounted for 60.8% (n=166) and 39.2% (n=107), respectively. The median survival duration was 26.4 months (range, 1.47–241.6 months).
Full table
TMEM213 is a prognostic gene that is associated with drug efficacy
Based on the TCGA database of 273 patients, 1,743 genes were screened out of 17,165 genes using Max stat package according to the following procedure. In the area between the 10th and 90th binary value, the selected cut point can obtain the optimal log-rank test (P<0.05). Then, based on the above 1,743 genes, univariate Cox analysis and KM curve (log-rank test) were used to screen out 297 genes with prognostic value.
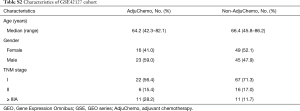
For these 297 genes, STEPP analysis was performed in GSE42127 (Table S1), and its main clinical information is listed in Table S2. The results showed that in the TMEM family, as TMEM213 expression increased, the OS rate at 3 years significantly increased in the ACT group compared with the non-ACT group (P=0.044). The difference in OS rate at 3 years was also statistically significant (P=0.044). Although the hazard ratio for OS did not achieve statistical significance, we did observe a better survival rate (P=0.137) (Figure 2 A,B,C).
Full table
Full table

Association of TMEM213 expression with clinicopathological variables
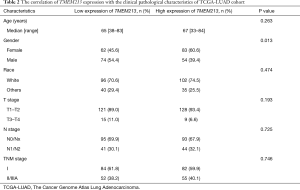
The correlation between clinical pathological parameters and TMEM213 expression in the TCGA database is shown in Table 2. There was no significant difference among patients with different TMEM213 expression levels while controlling for patients’ age, size, lymph node involvement, and TNM stage. Nevertheless, TMEM213 expression was statistically significantly associated with gender (P=0.013). High TMEM213 expression was more commonly seen among females.
Full table
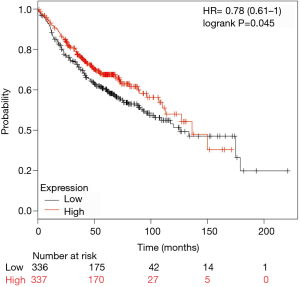
Influence of TMEM213 expression on survival and external validation
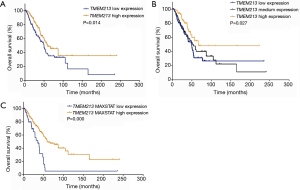
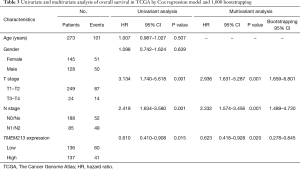
In order to assess the relationship between TMEM213 expression and prognosis with lung adenocarcinoma in TCGA database, Kaplan-Meier analysis showed patients with increased TMEM213 expression had longer OS (log-rank P=0.014, 0.027, and 0.000 for dichotomous, trichotomous and maximally selected log-rank modeling of TMEM213 expression, respectively) (Figures 3,S1). Univariate analysis using Cox PH models showed that lower T stage, lower N stage, and higher TMEM213 expression significantly predicted prolonged OS (P=0.001, 0.001, and 0.015, respectively). In the multivariable Cox PH analysis, TMEM213 expression [hazard ratio (HR) =0.623, P=0.020], T stage (HR =2.936, P=0.001), and N stage (HR =2.332, P=0.001) were associated with prolonged OS, and this prognostic model for lung adenocarcinoma was further validated using 1,000 bootstrapping replications (Table 3).
Full table
For external validation, patients with high expression of TMEM213 showed significantly longer OS than those with low expression level (log-rank P=0.045) in the online Kaplan-Meier plotter database (Figure 4).
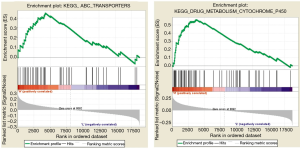
GSEA analysis for TMEM213 expression in lung adenocarcinoma
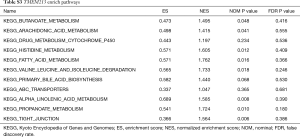
To investigate the possible function and mechanism of TMEM213 in lung adenocarcinoma, the enriched pathway was analyzed (Figure 5). High-throughput RNA sequencing data from the lung adenocarcinoma group in TCGA and GSE42127 were used for GSEA. The results showed that the expression of TMEM213 was related to the metabolic pathway of P450, ABC transporter, butyric acid, arachidonic acid, tryptophan, fatty acid, histidine, and bile acid synthesis and other metabolic pathways (Tables 4,S3).
Full table
Full table
Discussion
Several studies have shown that prognostic gene signatures can also be predictive of benefit from ACT, but few of them tested cohorts using adjuvant paclitaxel-carboplatin treatments (21,25,26). The NATCH trial failed to demonstrate patients who underwent resection could benefit from adjuvant paclitaxel-carboplatin. This may well be due to the fact that most patients in this study are stage I disease who are not known to benefit from chemotherapy (7). The Cancer and Leukemia Group B 9633 trial, comparing adjuvant paclitaxel-carboplatin with observation in patients with stage IB disease, was not powered to address improvement in OS with chemotherapy. Nonetheless, it could be considered as a treatment option in selected stage IB patients who had tumors ≥4.0 cm in diameter (27). In this study, we tried to identify a possible target in the TMEM family and elucidate its potential as a prognostic biomarker for the benefit from carboplatin plus taxane-based ACT in resected lung adenocarcinoma patients.
Bioinformatics analysis is a well-established method which utilizes online expression profile datasets and has been widely used to help researchers identify potential biomarkers. We analyzed the expression profile of lung adenocarcinoma in TCGA and screened a microarray dataset of GSE42127 by STEPP. We found that TMEM213 expression was significantly correlated with gender, and survival analysis showed that patients with high TMEM213 expression had significantly better OS than those with low TMEM213 expression. Univariate and multivariate Cox analysis indicated that TMEM213 was an independent predictor for OS improvement. The STEPP approach was developed to allow researchers to investigate the heterogeneity of treatment effects on survival outcomes across values of a (continuously measured) covariate, such as TMEM213 expression (22). Our study showed that with the increase of TMEM213 expression, the treatment group could benefit from adjuvant paclitaxel-carboplatin, and, as the value of gene expression increased, so did the survival benefit.
Our finding was reliable and generalizable for the following reasons. First, the cutoff value of TMEM213 expression, whether it was selected by the median method, the trisection method, or the most selective log-rank statistical analysis method, all showed that the patients with high expression had a good prognosis. Second, our Cox regression model was robust, and our findings were further confirmed by 1,000 internal bootstrap replications and external validation using an online high-throughput dataset.
Our findings have value for clinical application. We found that the prognostic value of TMEM213 signature was independent of T and N stage in the stepwise Cox regression. Presently, the TNM stage has been viewed as an important factor for predicting survival, but clinically, we can observe that patients with the same TNM stage might have a different prognosis. This highlights the importance of identifying more personalized biomarkers for a more precise survival prediction of patients with lung adenocarcinoma. Additionally, most current molecular signatures for lung cancer are prognostic only and do not provide any estimation as to whether a patient would benefit from ACT. STEPP results suggest that the expression of TMEM213 can guide our future selection and predictive efficacy of postoperative ACT in patients with radical lung adenocarcinoma. However, this trend still needs further validation in perspective, well-balanced studies with large sample size.
The characteristics of TMEM213 involving lung adenocarcinoma have not been reported in any functional studies. We speculate that it might be related to the growth of lung cancer. Alveolar type II cells can secrete surfactant, play an important role in ion transport, and contribute to the regulation of alveolar fluid and epithelial repair after lung injury (28). Lin found that lung adenocarcinoma can initiate in alveolar type II cells (29). One study found that TMEM213 expression was significantly upregulated in alveolar type II cells under a hypoxic environment induced by submerged condition (30). Yu found that long-term exposure to hypoxia repressed tumor progression of the lung cancer from A549 cells (31), and thus presumed that the high expression of TMEM213 gene may have the effect of inhibiting lung cancer. In addition, it is generally believed that TMEM213 is located in the endoplasmic reticulum (the reliability is 44.4%), and it has an endoplasmic relocation signal (9,32). Endoplasmic reticulum stress has a significant effect on the proliferation and growth of almost all types of cancer, including lung cancer (32). Hung found that dehydrocostuslactone (DHE) caused the apoptosis of NSCLC cells via the stress response of the endoplasmic reticulum (33). Li indicated that chaetocin induced the stress response signal of the endoplasmic reticulum and led to death receptor-5 dependent apoptosis in NSCLC (34). Thus, it is important to investigate the functional roles of TMEM213.
Enriched pathway analysis revealed the high TMEM213-expression-enriched gene signature of “DRUG_METABOLISM_CYTOCHROME_P450” and “ABC_TRANSPORTERS” from the KEGG pathway database. Both gene sets were associated with clinical drug resistance of paclitaxel. Paclitaxel is a potent anti-cancer agent that binds to β-tubulin and prevents mitosis through microtubule overstabilization. It is an effective chemotherapeutic agent to treat ovarian, breast, gastric, and lung cancers, and the response rate of paclitaxel-based chemotherapy for lung cancer is 30–40% (35). The P450 enzymes in tumor cells may inactivate the anticancer drug, and the overexpression of the ABC transporters may encode the efflux pump, which leads the drug to be discharged, thereby hindering the retention of the drug in the cell, which can have a negative effect on chemotherapeutic-mediated tumor cell death. Recent studies in vitro have shown that paclitaxel treatment increases the level of P450 in human hepatocytes as well as ABC transporters in colon tumor cells, and thus may influence their own metabolism and elimination (36). It has also been reported that a weekly paclitaxel chemotherapy regimen can induce P450 enzyme and ABC protein expression. This can maintain the plasma drug concentration of paclitaxel targeting the proliferating endothelial cells within the tumor as opposed to neoplastic cells themselves so that the role of paclitaxel chemotherapy is more effective (37). We found that high expression of TMEM213 was positively correlated with P450 pathway and ABC transporter pathway; these might lead to pharmacokinetically relevant changes in drug metabolism and elimination such that a modification in the concentration of paclitaxel might be beneficial.
Several limitations to this study needed to be noted. First, this study analyzed the online data of lung adenocarcinoma on TCGA and GEO but did not use clinical samples to validate. Second, our study did not investigate the mechanism behind the predictive and prognostic value of TMEM213 in lung adenocarcinoma. Experimental studies on cancer cell lines and xenograft models will provide more information to understand its functional role better. Third, our conclusions need to be validated by prospective controlled trials with large sample sizes guided by a priori conducted power analysis.
Conclusions
In summary, this study provided the first evidence that TMEM213 presence in lung adenocarcinoma is related to gender. High expression of TMEM213 suggests a good prognosis. Survival analysis with online data supports the results above. Furthermore, we found that high TMEM213 expression is more likely to benefit from adjuvant paclitaxel-carboplatin. Taken together, TMEM213 may serve as a potential candidate biomarker and therapeutic target for lung adenocarcinoma. Future studies will focus on the verification of our findings in clinical practices and the functional roles of this innovative signature.
Acknowledgments
Funding: The work was supported by the National Key Research and Development Program of China (Grant No. 2016YFC1303800), the Planned Science and Technology Projects of Liaoning Province (No. 2014226033).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Selvaggi G, Scagliotti GV. Histologic subtype in NSCLC: does it matter? Oncology (Williston Park) 2009;23:1133-40. [PubMed]
- Hung JJ, Jeng WJ, Chou TY, et al. Prognostic value of the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification on death and recurrence in completely resected stage I lung adenocarcinoma. Ann Surg 2013;258:1079-86. [Crossref] [PubMed]
- Chang WJ, Sun JM, Lee JY, et al. A retrospective comparison of adjuvant chemotherapeutic regimens for non-small cell lung cancer (NSCLC): paclitaxel plus carboplatin versus vinorelbine plus cisplatin. Lung Cancer 2014;84:51-5. [Crossref] [PubMed]
- Sugaya M, Uramoto H, Uchiyama A, et al. Phase II trial of adjuvant chemotherapy with bi-weekly carboplatin plus paclitaxel in patients with completely resected non-small cell lung cancer. Anticancer Res 2010;30:3039-44. [PubMed]
- Yamashita Y, Kataoka K, Ishida T, et al. A feasibility study of postoperative adjuvant therapy of carboplatin and weekly paclitaxel for completely resected non-small cell lung cancer. J Thorac Oncol 2008;3:612-6. [Crossref] [PubMed]
- Felip E, Rosell R, Maestre JA, et al. Preoperative chemotherapy plus surgery versus surgery plus adjuvant chemotherapy versus surgery alone in early-stage non-small-cell lung cancer. J Clin Oncol 2010;28:3138-45. [Crossref] [PubMed]
- Wallerek S, Sorensen JB. Biomarkers for efficacy of adjuvant chemotherapy following complete resection in NSCLC stages I-IIIA. Eur Respir Rev 2015;24:340-55. [Crossref] [PubMed]
- Wrzesiński T, Szelag M, Cieslikowski WA, et al. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer 2015;15:518. [Crossref] [PubMed]
- Fuller CM. Time for TMEM? J Physiol 2012;590:5931-2. [Crossref] [PubMed]
- Zhou X, Popescu NC, Klein G, et al. The interferon-alpha responsive gene TMEM7 suppresses cell proliferation and is downregulated in human hepatocellular carcinoma. Cancer Genet Cytogenet 2007;177:6-15. [Crossref] [PubMed]
- Liu F, Cao QH, Lu DJ, et al. TMEM16A overexpression contributes to tumor invasion and poor prognosis of human gastric cancer through TGF-beta signaling. Oncotarget 2015;6:11585-99. [PubMed]
- Dobashi S, Katagiri T, Hirota E, et al. Involvement of TMEM22 overexpression in the growth of renal cell carcinoma cells. Oncol Rep 2009;21:305-12. [PubMed]
- Cuajungco MP, Podevin W, Valluri VK, et al. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem 2012;114:705-12. [Crossref] [PubMed]
- Yang S, Li H, Liu Y, et al. Elevated expression of MAC30 predicts lymph node metastasis and unfavorable prognosis in patients with epithelial ovarian cancer. Med Oncol 2013;30:324. [Crossref] [PubMed]
- Guo J, Chen L, Luo N, et al. Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol Rep 2015;33:3124-30. [Crossref] [PubMed]
- Cheng Z, Guo J, Chen L, et al. Overexpression of TMEM158 contributes to ovarian carcinogenesis. J Exp Clin Cancer Res 2015;34:75. [Crossref] [PubMed]
- Kamiński MJ, Kaminska M, Skorupa I, et al. In-silico identification of cardiovascular disease-related SNPs affecting predicted microRNA target sites. Pol Arch Med Wewn 2013;123:355-63. [PubMed]
- Ha SY, Sung J, Ju H, et al. Epstein-Barr virus-positive nodal peripheral T cell lymphomas: clinicopathologic and gene expression profiling study. Pathol Res Pract 2013;209:448-54. [Crossref] [PubMed]
- Petricoin EF, Hackett JL, Lesko LJ, et al. Medical applications of microarray technologies: a regulatory science perspective. Nature Genetics 2002;32:474-9. [Crossref] [PubMed]
- Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577-86. [Crossref] [PubMed]
- Yip WK, Bonetti M, Cole BF, et al. Subpopulation Treatment Effect Pattern Plot (STEPP) analysis for continuous, binary, and count outcomes. Clin Trials 2016;13:382-90. [Crossref] [PubMed]
- Bonetti M, Gelber RD. A graphical method to assess treatment-covariate interactions using the Cox model on subsets of the data. Stat Med 2000;19:2595-609. [Crossref] [PubMed]
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Zhu CQ, Ding K, Strumpf D, et al. Prognostic and Predictive Gene Signature for Adjuvant Chemotherapy in Resected Non–Small-Cell Lung Cancer. J Clin Oncol 2010;28:4417-24. [Crossref] [PubMed]
- Chen DT, Hsu YL, Fulp WJ, et al. Prognostic and Predictive Value of a Malignancy-Risk Gene Signature in Early-Stage Non–Small Cell Lung Cancer. J Natl Cancer Inst 2011;103:1859-70. [Crossref] [PubMed]
- Strauss GM, Herndon JE 2nd, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol 2008;26:5043-51. [Crossref] [PubMed]
- Mason RJ. Biology of alveolar type II cells. Respirology 2006;11 Suppl:S12-5. [Crossref] [PubMed]
- Lin C, Song H, Huang C, et al. Alveolar type II cells possess the capability of initiating lung tumor development. PLoS One 2012;7:e53817. [Crossref] [PubMed]
- Ito Y, Zemans R, Correll K, et al. Stanniocalcin-1 is induced by hypoxia inducible factor in rat alveolar epithelial cells. Biochem Biophys Res Commun 2014;452:1091-7. [Crossref] [PubMed]
- Yu L, Hales CA. Long-term exposure to hypoxia inhibits tumor progression of lung cancer in rats and mice. BMC Cancer 2011;11:331. [Crossref] [PubMed]
- Clarke HJ, Chambers JE, Liniker E, et al. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014;25:563-73. [Crossref] [PubMed]
- Hung JY, Hsu YL, Ni WC, et al. Oxidative and endoplasmic reticulum stress signaling are involved in dehydrocostuslactone-mediated apoptosis in human non-small cell lung cancer cells. Lung Cancer 2010;68:355-65. [Crossref] [PubMed]
- Liu X, Guo S, Liu X, et al. Chaetocin induces endoplasmic reticulum stress response and leads to death receptor 5-dependent apoptosis in human non-small cell lung cancer cells. Apoptosis 2015;20:1499-507. [Crossref] [PubMed]
- Pujol JL, Barlesi F, Daures JP. Should chemotherapy combinations for advanced non-small cell lung cancer be platinum-based? A meta-analysis of phase III randomized trials. Lung Cancer 2006;51:335-45. [Crossref] [PubMed]
- Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med 2001;7:584-90. [Crossref] [PubMed]
- Gustafson DL, Long ME, Bradshaw EL, et al. P450 induction alters paclitaxel pharmacokinetics and tissue distribution with multiple dosing. Cancer Chemother Pharmacol 2005;56:248-54. [Crossref] [PubMed]