Differentially expressed miRNAs in circulating exosomes between atrial fibrillation and sinus rhythm
Introduction
Atrial fibrillation (AF) is the commonest cardiac arrhythmia in clinics, leading to many complications like stroke and thromboembolic events (1). Approximately 10% of ischemic strokes are associated with AF first diagnosed at the time of stroke (2). AF increases the risk of death and disability, being a burden on the cost of health care. The molecular mechanisms of AF are complex, and many studies have revealed the key mechanisms like Ca2+-dependent signaling pathways (3), inflammatory and immune pathways causing electric remodeling and structural remodeling (4,5). Some studies revealed some cardiomyocyte-derived miRNAs targeting pathways involved in cardiomyocyte metabolism and remodeling (miR-223, miR-1, miR-21, miR-29b, miR-26, miR-208a, miR-328 and miR-499) (6,7), but few studies focus on the miRNAs transported by exosomes which may influence the AF pathophysiology. Tian et al. reported the levels of two upregulated miRNAs in lung cancer were significantly higher in exosomes than plasma (8), and Emanueli et al. found cardiac miRNAs in the whole plasma did not correlate significantly with cTn-I in patients underwent coronary artery-bypass-graft surgery, while cTn-I was positively correlated with the plasma exosome level and the exosomal cardiac miRNAs (9). So plasma-derived exosomal miRNAs may play a better role than plasma miRNAs in diagnosis or prognosis. Some studies reported therapeutic bioactivity on heart of exosomes secreted by human embryonic stem cells and mesenchymal stem cells (10,11). Identifying the exosomal difference may also contribute to providing new biomarkers or even exosomal therapeutic methods for AF.
Exosomes are small (30–150 nm) membrane vesicles released into the extracellular environment after the multivesicular bodies fuse with the plasma membrane (12). miRNAs are small non-coding RNAs (ncRNAs) which regulate gene expression (13). Exosomes are enriched with selected RNAs from parent cells (14). Exosomal miRNAs in particular can potently and fundamentally alter the transcriptome of recipient cells (15) and serve as diagnostic or prognostic biomarkers in many diseases (16,17).
Liquid biopsies have advanced rapidly recently for diagnostic and prognostic application (18). Considering this, we are motivated to conduct a transcriptomic study using plasma samples from non-valvular persistent AF patients and sinus rhythm (SR) patients. The goal of this study is to identify the exosomal miRNAs that may take effect in the initiation or maintenance of AF. With RNA-Sequencing (RNA-Seq) technology, we found 2,371 miRNAs, of which 39 are differentially expressed (DE) (including 21 upregulated miRNAs and 18 downregulated miRNAs) in non-valvular AF patients comparing to SR patients. Target genes of the DE miRNAs were predicted and put into gene ontology analysis and pathway analysis. Some of the DE miRNAs were validated by qRT-RCT. Overall, our study uncovered signaling pathways and exosomal miRNAs which are related with AF.
Methods
Patient recruitment and sampling
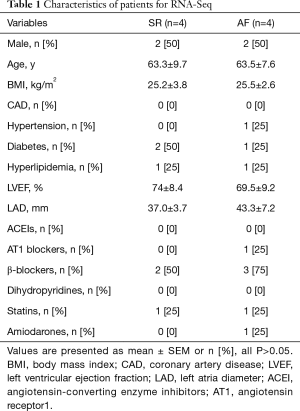
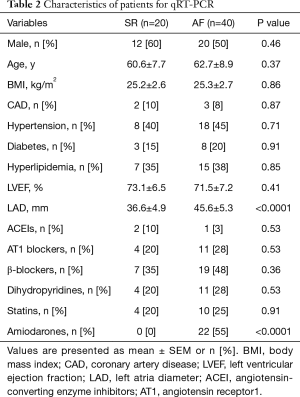
With informed consent signed, patients with non-valvular persistent AF and patients with SR who were admitted to Shanghai Changzheng Hospital from March 2016 to July 2018 were recruited. AF was defined according to electrocardiogram showing persistent AF more than 7 days. SR patients were admitted for chest distress without arrythmia. Those with valvular disease, patients with history of cardiac surgery or thyroid disease, those with hepatic or renal dysfunction or malignant tumor were excluded. For the small RNA-Sequencing phase, we used the blood sample of persistent AF patients (n=4) and patients with SR control (n=4). For qRT-PCR phase, persistent AF (n=40) and patients with SR control (n=20) were recruited. Whole blood samples were obtained before breakfast. Plasma was extracted from whole blood by centrifugation at 3,000 rpm for 10 min and stored in liquid nitrogen before exosome extraction. The study was conducted in accordance with the Declaration of Helsinki, under protocols approved by the Committee on Ethics of Biomedicine of the second Military Medical University. Characteristics of the patients are listed in Tables 1 and 2.
Full table
Full table
Isolation of Exosome and extraction of exosomal RNA
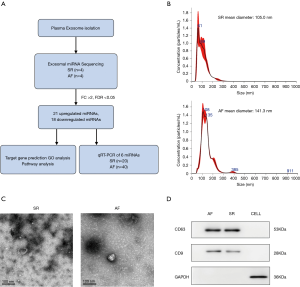
To remove cells and debris, the plasma was centrifugated at 10,000 ×g at 4 °C for 30 min, followed by twice of centrifugation at 100,000 ×g at 4 °C for 70 min, washing with PBS. The final precipitates containing exosomes was re-suspended in PBS. With Nanosight LM10 System (Malvern Panalytical Ltd) and the particle-tracking and fast video capture software, the size distribution of exosomes and nanoparticle concentrations was analyzed. Exosomes were examined by transmission electron microscopy (FEI Quanta 250) after being suspended in uranyl acetate, dropped on copper grids, precipitated, dried and imaged. These exosomal markers CD63, CD9 and GAPDH expression level were assessed by Western blotting.
Western blot analysis
Total proteins were extracted from exosome and Hela cells using lysis buffer. Total protein was loaded a 12% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and then transferred to a polyvinylidene fluoride membrane. The membrane was incubated by primary antibodies anti-CD63 (1:1,000, SBI, MA, USA), anti-CD9 (1:1,000, SBI, MA, USA), GAPDH (1:1,000, Beyotime, China) followed by relevant secondary antibodies joined and visualized by an electro-chemiluminescence (ECL) reagent.
Small RNA library establishment and miRNA sequencing
Trizol reagent (Invitrogen, CA, USA) was applied to isolate total RNA from exosome. With Nanodrop (Thermo Fisher Scientific, MA, USA) and 1% gel electrophoresis, the amount and purity of RNA were evaluated. Each purified RNA sample was put into adapter ligation, cDNA synthesis, PCR amplification and RNA libraries construction with the miRNA Library Prep Kit (New England Biolabs, MA, USA) according to the Illumina protocol. Besides, RNA sequencing was performed on Hiseq 2500 (Illumina, CA, USA). We generated datasets with 50 million reads sequencing depth of the multiplexed samples. And the raw data were refined with FastQC to filter out short (<15 nt) and low-quality reads. The high-quality reads were matched to miRBase (http://www.mirbase.org/) database to reveal known miRNAs.
qRT-PCR validation
Total RNA was reverse-transcribed into cDNA using the miRcute Plus miRNA First-Strand cDNA Synthesis Kit (#KR211-02, Tiangen Biotech Co., Ltd, Beijing, China) and then measured by miRcute Plus miRNA qPCR Detection Kit (SYBR Green) (#FP411-02, Tiangen Biotech Co., Ltd, Beijing, China) with Qiagen predesigned primers. All kits were used according to the manufacturer’s instructions. The expression levels of the miRNAs were calculated with the 2−△△Ct method (19), with U6 acting as the internal reference.
Statistical analysis
DESeq2.0 algorithm is used to analyze differential expression of miRNAs, and statistical significance was reported if fold change (FC) >2 or FC <1/2, false discovery rate (FDR) <0.05. Measurement data are presented as mean ± SEM and compared using student’s t-test when appropriate and were considered statistically significant at P<0.05. Categorical data are presented as n (%) and compared using Chi-square test or Fisher’s exact method. Univariate and multivariate logistic regression analysis were used to evaluate the relation between miRNAs and AF.
Results
Identification of exosome
The isolated exosomes were analyzed by Nanosight LM10 System, transmission electron microscopy and Western blotting (Figure 1). Nanoparticles with diameter of 30–150 nm are regarded as exosomes, and no significant difference of exosome concentration was found between AF and SR. Both AF and SR plasma exosomes are positive for exosomal markers, CD63 and CD9, and negative for GAPDH.
Exosomal miRNA-Seq of plasma sample
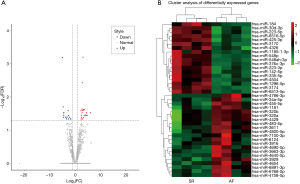
The filtered data of exosomal small RNAs was compared to miRbase database to obtain the alignment result, and the length distribution of the reads and the base preference were compared. Total read counts of AF and SR samples were normalized and compared in total online: http://fp.amegroups.cn/cms/5d5bb36ab3c55ece8b6fe3d97cb60dde/JTD.2019.09.50-1.xls. The detailed expression level and FC of the 39 DE miRNAs were listed in total online: http://fp.amegroups.cn/cms/d7cdc000d7326a48cfcccc7aa43493c6/JTD.2019.09.50-2.xls. The results were visualized in a volcano plot where the x-axis is the log2 ratio of the fold-change (AF/SR) and the y-axis is the –log10 ratio of FDR (Figure 2). With the DESeq2.0 algorithm, we found 21 upregulated miRNAs and 18 downregulated miRNAs (Table 3 and colorful points shown in volcano plot in Figure 2). A heat-map was drawn according to the DE miRNAs (Figure 2).
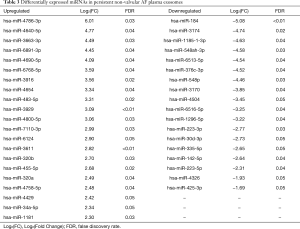
Full table
Studies about the 39 DE miRNAs were searched on PubMed. Of these, only 8 (has-miRNA-223-3p, has-miRNA-223-5p, has-miRNA-320a, has-miRNA-320b, has-miRNA-34a-5p, has-miRNA-184, has-miRNA-142-5p, has-miRNA-483-5p) were reported to play a role in cardiac diseases.
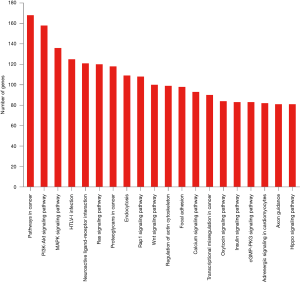
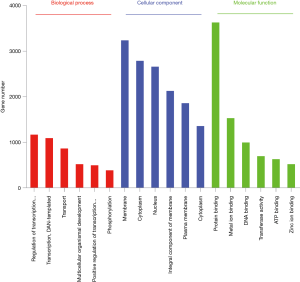
Target genes were predicted by miranda (score >150, energy <−20) and RNAhybrid (energy <−25) for the significantly different miRNAs, and the intersection of the two algorithms was used as the final forecast result of target gene in total online: http://fp.amegroups.cn/cms/0d09f67fb86ebf5c5fc7720a3c4214a0/JTD.2019.09.50-3.xls. The target genes underwent gene ontology (GO) analysis to better understand these transcripts using Fisher’s exact test, the difference was considered significant at P<0.05. The target genes were classified according to their “Biological Process”, “Molecular Function”, and “Cellular Component” in total online: http://fp.amegroups.cn/cms/f16328e08e3dcfa4b195f0041c4f692d/JTD.2019.09.50-4.zip. Top 6 of each kind of GO enrichment is listed in Figure 3. The top 3 types of “Biological Process” were “regulation of transcription, DNA-templated”, “transcription, DNA-templated”, “transport”, and the top 3 types of “Cellular Component” were “membrane”, “cytoplasm”, “nucleus”, while the top 3 types of “Molecular Function” were “protein binding”, “metal ion binding”, “DNA binding”. The target genes were also performed pathway analysis using KEGG (http://www.genome.jp/Kegg/) and Fisher’s exact method, significant difference was defined as P<0.05 in total online: http://fp.amegroups.cn/cms/3816dffd1eb444e66c9fc66f37462f62/JTD.2019.09.50-5.xlsx. Some of the pathways are related with myocardial remodeling (PI3K-Akt signaling pathway, adrenergic signaling in cardiomyocytes, focal adhesion, Wnt signaling pathway, calcium signaling pathway) and oxidative stress (MAPK signaling pathway, oxytocin signaling pathway) (Figure 4).
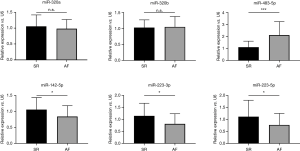
qRT-PCR validation of DE exosomal miRNAs
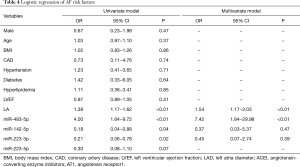
We performed qRT-PCR on 6 of the DE exosomal miRNAs (miR-320a, miR-320b, miR-483-5p, miR-142-5p, miR-223-3p, miR-223-5p) that are related with heart diseases and highly expressed in human tissue or blood reported in miRbase and have high signals in our miRNA-Seq results (Figure 5). The miR-483-5p (2.11±1.13 vs. 1.11±0.50, P<0.001) expression in AF patients is significantly higher than SR patients, and the expression of miR-142-5p (0.84±0.34 vs. 1.06±0.37, P=0.026), miR-223-3p (0.81±0.43 vs. 1.14±0.53, P=0.011), miR-223-5p (0.77±0.48 vs. 1.11±0.69, P=0.028) are lower in AF patients. Univariate logistic analysis shows miR-483-5p, miR-142-5p, miR-223-3p are related with AF, while multivariate logistic analysis suggests miR-483-5p is independently in correlation with AF (Table 4).
Full table
Discussion
As far as we know, this is the first transcriptomic study in patients with non-valvular persistent AF performing exosomal miRNA-sequencing technology and identifying the DE exosomal miRNAs. With the advanced high-throughput miRNA-Seq, we revealed 39 miRNAs that were DE in patients with non-valvular AF. We validated 4 of these DE transcripts using qRT-PCR, indicating these miRNAs may take effect through transportation by exosomes in AF pathogenesis. Another study performed by Joung’s group compared expression of miRNA in serum exosomes between persistent AF patients and supraventricular tachycardia patients and found 5 miRNAs (miR-103a, miR-107, miR-320d, miRNA-486, let-7b) with elevated expression in AF patients (20). They used microarray analysis, which is dependent upon the signals generated via hybridization. While the high-throughput miRNA-Seq we used shows the expressing levels of the miRNAs directly and possesses higher sensitivity. The pathway enrichment of the target genes of DE exosomal miRNAs revealed that the functions of the targeted genes were related with myocardial electrical and structural remodeling, fibrosis and oxidative stress.
As for the DE miRNAs, Harling et al. compared the miRNAs in right atrial biopsies between post-operative AF and SR patients undergoing non-emergent, on-pump CABG, revealing 11 upregulated miRNAs and 5 downregulated miRNAs, of which only 1 miRNA (hsa-miR-483-5p) was also upregulated in our study, this study also suggests that higher serum miRNA 483-5p may serve as a predictor of post-operative AF (21). Nicola Cooley et al. found 15 upregulated miRNAs and 32 downregulated miRNAs of right atria between valvular heart disease patients with AF and SR. Only 2 of these miRNAs (miR-483-5p, miR-142-5p) are also differently expressed in our study. miR-483-5p was upregulated in both studies, while miR-142-5p was upregulated in Nicola Cooley’s study and downregulated in our study (22). This may reflect the different mechanism between valvular AF and non-valvular AF. And this further showed the advantage of our study that we recruited non-valvular AF patients and excluded complex confounding factors like rheumatic valvular heart disease and degenerative valvular heart disease. In another study, serum miR-19b, miR-223 and miR-483-5p showed higher diagnostic accuracy than Point of care troponin assay and could even predict acute myocardial infarction in patients with negative troponin in early phase (23). miR-483-5p, part of the intron of insulin-like growth factor 2 (IGF-2), plays a role in inhibiting angiogenesis endogenously by targeting serum response factor and causing defective vascular morphogenesis (24). miR-483-5p also targets suppressor of cytokine signaling-3 (SOC-3) and correlated with insulin resistance, obesity and independently correlated with de novo diabetes or cardiovascular disease (25). But the detailed underlying mechanism between miR-483-5p and AF has not been elucidated. Zhan et al. reported overexpression of miR-142-5p caused extensive cell injury and apoptosis of hypoxia-exposed H9C2 cardiomyoblasts and primary cardiomyocytes through downregulating Sirtuin7 (SIRT7) (26). While Wang A and colleagues found lower circulating miR-142-5p was significantly correlated with chronic heart failure (27). The group of Kun Wang demonstrated that miR-223 (miR-223-3p) serves as a positively regulator of myocardial hypertrophy and heart failure through targeting apoptosis repressor with a caspase recruitment domain (ARC) (28). Whereas Qin et al. displayed that miR-223-3p which targets NOD-like receptor protein 3 (NLRP3) and inhibitor of κB (IκB) kinase α, and miR-223-5p which targets tumor necrosis factor receptor1 (TNFR1) and death receptor 6 (DR6), synergistically suppressed inflammation and necroptosis of ischemia/reperfusion treated cardiomyocyte (29). The functional roles of these miRNAs underlying AF are yet to be studied in vitro and in vivo.
Previous studies on AF focus on the miRNAs in plasma or tissue, and this is the first study revealing the plasma exosomal miRNAs differences between non-valvular persistent AF patients and SR patients. As exosomes are highly conserved in Eukaryota taxa and play important even central roles in disease (30), they may mediate the benefits of embryonic or mesenchymal stem cell therapy and may be therapeutic candidates supplanting the need for cell transplantation (14,31). Exosomal miRNAs constitute approximately 10–15% of serum miRNAs and act as intercellular messengers and they are protected from degradation by exosomes (32). As they stay more stable in the exosomes than other part of the blood, measuring exosomal clinical biomarkers can obtain higher sensitivity than total biological fluids.
Bang et al. identified miR-21-3p enriched in fibroblast exosomes as a potent pathogenic molecule which can result in myocardial hypertrophy. Their later studies revealed the mechanism and indicated that after fibroblast exosomes are taken up by cardiomyocytes via endocytosis, miR-21-3p silences sorbin, SH3 domain-containing protein 2 (SORBS2) and PDZ and LIM domain 5 (PDLIM5), leading to hypertrophy and antagonism of miR-21-3p can attenuate the hypertrophy of mice model with Ang II-induced cardiac hypertrophy (33). These outcomes indicate that researches on exosome-mediated communication systems can lead to findings of novel mechanisms resulting in myocardial remodeling or cardiac failure. Other exosomal miRNAs suggest protective function, such as miR-425 and miR-744 suppress cardiac fibrosis by negatively regulating TGFβ1, presenting the potential of being a predictor of cardiac fibrosis and heart failure (34). Specific miRNAs are selectively packaged into exosomes or other carriers that are released into the blood stream during disease conditions and they are regulated differently between plasma and plasma-derived exosomes (35). Exosomes are naturally suitable for the transportation and cell-to-cell delivery of proteins and small nucleic acids. This trait makes them highly attractive as pharmaceutical ingredient. Furthermore, their biophysical properties make it easy to extract exosomes and manipulate the protein and RNA contents (36). Thus, in addition to being regarded as therapeutic agents themselves, natural exosomes and exosome-mimetic vesicles are deemed to be potentially therapeutic Trojan Horses to transport biological drugs, including small RNAs and recombinant proteins, which escape from degradation by enzymes in the blood. Exosomes effectively deliver agents across different biological barriers and Alvarez-Erviti et al. firstly modified exosomes to transport exogenous short interfering RNA targeting mice brain, showing their special features for future neurodegenerative diseases therapy application (37). Considering exosomal miRNAs may present protective impact on cardiomyocytes, and exosomes are promising tools for antiarrhythmic miRNAs delivery, providing a novel treatment for arrhythmia. Some exosomal miRNAs have been studied in new therapeutic strategy of AF. As vitro overexpression of miR-1 or miR-133 alleviates cardiac hypertrophy (38), Park et al. loaded exosomes with miR-1 and miR-133a with electrophoresis and transfection method and found these modified exosomes protect HL-1 from electric remodeling, contractile dysfunction, autophagy and depolymerization of microtubules induced by tachypacing (39). Although exosomes display attractive possibilities for cardiovascular treatment, these novel therapies remain unexplored field which we are committed to exploit.
Conclusions
In summary, we found no statistical difference in the concentration of exosomes between AF and SR plasma, but we discovered 39 differently expressed exosomal miRNAs and validated 4 of them (miR-483-5p, miR-142-5p, miR-223-3p, miR-223-5p) through qRT-PCR. These findings open up a new perspective in the complicated mechanism underlying cardiac fibrosis and AF in which the exosomes may play an important role. The exosomes carrying disease-resistant miRNAs or other agents are promising novel pharmacological intervention for AF. And the DE exosomal miRNAs exert enormous potential of acting as biomarkers in assessing severity or prognostic of AF and help selecting therapeutic strategy.
Acknowledgments
Funding: This study was funded by National Nature Science Foundation of China (No. 81670299, 81770244), Shanghai Science and Technology Committee Medicine Leading Project (No. 15411960100), Shanghai Science and Technology Commission (17ZR1439100) and Shanghai Leading Talent Program (No. 2015044).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki, under protocols approved by the Committee on Ethics of Biomedicine of the Second Military Medical University.
References
- Lam A, Goulouti E, Roten L. The search for atrial fibrillation and its impact on public health. Swiss Med Wkly 2017;147:w14447. [PubMed]
- Freedman B, Camm J, Calkins H, et al. Screening for Atrial Fibrillation: A Report of the AF-SCREEN International Collaboration. Circulation 2017;135:1851-67. [Crossref] [PubMed]
- Fan X, Yu Y, Lan H, et al. Ca2+/Calmodulin-Dependent Protein Kinase II (CaMKII) Increases Small-Conductance Ca2+-Activated K+ Current in Patients with Chronic Atrial Fibrillation. Med Sci Monit 2018;24:3011-23. [Crossref] [PubMed]
- Kim N, Jung Y, Nam M, et al. Angiotensin II affects inflammation mechanisms via AMPK-related signalling pathways in HL-1 atrial myocytes. Sci Rep 2017;7:10328. [Crossref] [PubMed]
- Liu L, Zheng Q, Lee J, et al. PD-1/PD-L1 expression on CD(4+) T cells and myeloid DCs correlates with the immune pathogenesis of atrial fibrillation. J Cell Mol Med 2015;19:1223-33. [Crossref] [PubMed]
- Clauss S, Sinner MF, Kaab S, et al. The Role of MicroRNAs in Antiarrhythmic Therapy for Atrial Fibrillation. Arrhythm Electrophysiol Rev 2015;4:146-55. [Crossref] [PubMed]
- Zhao ZH, Luo J, Li HX, et al. SOX2-mediated inhibition of miR-223 contributes to STIM1 activation in phenylephrine-induced hypertrophic cardiomyocytes. Mol Cell Biochem 2018;443:47-56. [Crossref] [PubMed]
- Tian F, Shen Y, Chen Z, et al. No Significant Difference between Plasma miRNAs and Plasma-Derived Exosomal miRNAs from Healthy People. Biomed Res Int 2017;2017:1304816. [Crossref] [PubMed]
- Emanueli C, Shearn AI, Laftah A, et al. Coronary Artery-Bypass-Graft Surgery Increases the Plasma Concentration of Exosomes Carrying a Cargo of Cardiac MicroRNAs: An Example of Exosome Trafficking Out of the Human Heart with Potential for Cardiac Biomarker Discovery. PLoS One 2016;11:e0154274. [Crossref] [PubMed]
- Lai RC, Arslan F, Lee MM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res 2010;4:214-22. [Crossref] [PubMed]
- Arslan F, Lai RC, Smeets MB, et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res 2013;10:301-12. [Crossref] [PubMed]
- Li P, Kaslan M, Lee SH, et al. Progress in Exosome Isolation Techniques. Theranostics 2017;7:789-804. [Crossref] [PubMed]
- Bernardo BC, Ooi JY, Lin RC, et al. miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. Future Med Chem 2015;7:1771-92. [Crossref] [PubMed]
- Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470-6. [Crossref] [PubMed]
- Ibrahim A, Marban E. Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology. Annu Rev Physiol 2016;78:67-83. [Crossref] [PubMed]
- Saadatpour L, Fadaee E, Fadaei S, et al. Glioblastoma: exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther 2016;23:415-8. [Crossref] [PubMed]
- Matsumoto Y, Kano M, Akutsu Y, et al. Quantification of plasma exosome is a potential prognostic marker for esophageal squamous cell carcinoma. Oncol Rep 2016;36:2535-43. [Crossref] [PubMed]
- Mader S, Pantel K. Liquid Biopsy: Current Status and Future Perspectives. Oncol Res Treat 2017;40:404-8. [Crossref] [PubMed]
- Zhong Y, Lin Y, Shen S, et al. Expression of ALDH1 in breast invasive ductal carcinoma: an independent predictor of early tumor relapse. Cancer Cell Int 2013;13:60. [Crossref] [PubMed]
- Mun D, Kim H, Kang JY, et al. Expression of miRNAs in circulating exosomes derived from patients with persistent atrial fibrillation. FASEB J 2019;33:5979-89. [Crossref] [PubMed]
- Harling L, Lambert J, Ashrafian H, et al. Elevated serum microRNA 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur J Cardiothorac Surg 2017;51:73-8. [Crossref] [PubMed]
- Cooley N, Cowley MJ, Lin RC, et al. Influence of atrial fibrillation on microRNA expression profiles in left and right atria from patients with valvular heart disease. Physiol Genomics 2012;44:211-9. [Crossref] [PubMed]
- Li L, Li S, Wu M, et al. Early diagnostic value of circulating microRNAs in patients with suspected acute myocardial infarction. J Cell Physiol 2019;234:13649-58. [Crossref] [PubMed]
- Qiao Y, Ma N, Wang X, et al. MiR-483-5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett 2011;585:3095-100. [Crossref] [PubMed]
- Gallo W, Esguerra JLS, Eliasson L, et al. miR-483-5p associates with obesity and insulin resistance and independently associates with new onset diabetes mellitus and cardiovascular disease. PLoS One 2018;13:e0206974. [Crossref] [PubMed]
- Zhan L, Lei S, Li W, et al. Suppression of microRNA-142-5p attenuates hypoxia-induced apoptosis through targeting SIRT7. Biomed Pharmacother 2017;94:394-401. [Crossref] [PubMed]
- Wang A, Kwee LC, Grass E, et al. Whole blood sequencing reveals circulating microRNA associations with high-risk traits in non-ST-segment elevation acute coronary syndrome. Atherosclerosis 2017;261:19-25. [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Qin D, Wang X, Li Y, et al. MicroRNA-223-5p and -3p Cooperatively Suppress Necroptosis in Ischemic/Reperfused Hearts. J Biol Chem 2016;291:20247-59. [Crossref] [PubMed]
- Lopez-Verrilli MA, Court FA. Exosomes: mediators of communication in eukaryotes. Biol Res 2013;46:5-11. [Crossref] [PubMed]
- Bei Y, Chen T, Banciu DD, et al. Circulating Exosomes in Cardiovascular Diseases. Adv Exp Med Biol 2017;998:255-69. [Crossref] [PubMed]
- Siegel SR, Mackenzie J, Chaplin G, et al. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep 2012;39:6219-25. [Crossref] [PubMed]
- Bang C, Batkai S, Dangwal S, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest 2014;124:2136-46. [Crossref] [PubMed]
- Wang L, Liu J, Xu B, et al. Reduced exosome miR-425 and miR-744 in the plasma represents the progression of fibrosis and heart failure. Kaohsiung J Med Sci 2018;34:626-33. [Crossref] [PubMed]
- Xie JX, Fan X, Drummond CA, et al. MicroRNA profiling in kidney disease: Plasma versus plasma-derived exosomes. Gene 2017;627:1-8. [Crossref] [PubMed]
- El-Andaloussi S, Lee Y, Lakhal-Littleton S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc 2012;7:2112-26. [Crossref] [PubMed]
- Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 2011;29:341-5. [Crossref] [PubMed]
- Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613-8. [Crossref] [PubMed]
- Park H, Park HL, Kim HE, et al. Exosome derived from atrial fibrillation patients prevents proarrhythmic remodeling by suppressing autophagy in pacing induced tachycardia model Eur Heart J 2017;38:1562. [Crossref]