
Molecular epidemiology of respiratory viruses among Malaysian Young children with a confirmed respiratory infection during 2014–2015
Introduction
In many low- and middle-income countries, acute respiratory tract infections (ARTIs) are the leading cause of morbidity and mortality among children less than 5 years of age (1,2). ARTIs commonly affect children between 2 to 24 months old, with the highest incidence of infection afflicting babies age between 3 and 6 months (3). In 2000, an estimated 1.9 million children succumbed to ARTIs globally, with 70% of the death occurred in Southeast Asian and African regions (1). This finding was supported by a systemic review in which approximately 1.6 million children worldwide perished due to ARTIs (4).
ARTIs are commonly caused by viruses; such as respiratory syncytial virus (RSV), adenovirus, influenza virus, parainfluenza virus, human bocavirus, and human coronavirus (5-10). However, there is a scarcity on information regarding these respiratory infections especially in developing countries such as Malaysia. In these countries, majority of the paediatric patients who presented with ARTIs are often treated symptomatically in outpatient clinics. Due to the similarity in clinical symptoms at presentation, it is a challenge to identify the underlying viruses leading to the ARTIs. A retrospective study showed that 1–2% of US infants presenting with lower respiratory tract infection were hospitalized (11).
There is limited epidemiological data available on viral ARTIs in Malaysia. Many of the published data were from more than 10 years ago (12-14). In a retrospective study, many of the common respiratory viruses such as human metapneumovirus, coronavirus HKU1, human bocavirus, and influenza A (H1N1) virus were not included due to the lack of virological testing methods (15). Viral culture is a promising method for the detection of respiratory viral pathogens. However, this method required up to 14 days to confirm the detection (16). Quick test can be reported on the same day using immunofluorescence staining. However, its sensitivity is low (16,17). The development of multiplex polymerase chain reaction (PCR) could detect multiple pathogens in a single sample. This allows for rapid and cost-effective diagnosis of ARTIs.
The study aims to determine the aetiologies of viral ARTIs among young children in Malaysia. A comprehensive and up-to-date epidemiological data on viral ARTIs would be vital for clinical management especially in preventing unnecessary antibiotic usage and hospitalization.
Methods
Study design
This is a prospective surveillance study conducted between July 2014 and July 2015. All children aged 0–5 years old with ARTIs who were seen by pediatricians in Kuala Lumpur hospitals had their respiratory samples collected and sent for analysis (n=394). ARTIs included both upper or lower respiratory tract infections. Patients with symptom onset of more than 14 day were excluded. Patients who were found to have more than 1 positive specimens within the same week were excluded from the final analysis. All other samples obtained from the patients with ARTIs were included in the analysis. As the specimen might not have been taken at the point of admission, we were unable to fully exclude ARTIs of nosocomial origin. Using flocked swabs, nasopharyngeal swabs were taken from the patients. The swabs were then transported in 3.0 mL of viral transport medium (VTM) via cold chain to the virology laboratory. Upon receiving, the laboratory technicians would process and test the samples immediately.
Viral identification
In this study, the detection of RSV, influenza A (with additional subtyping of H1, H3, and H1N1), influenza B, parainfluenza virus types 1–4, adenovirus, human enterovirus/human rhinovirus, human bocavirus, human metapneumovirus, human coronavirus 229E, NL63, OC43, and HKU1 subtypes was made using the Luminex xTAG® Respiratory viral panel (RVP) Fast v2 assay. Using Qiagen MinElute Virus Spin Kit (Qiagen, Germany), the viral nucleic acids were extracted from VTM as per the instructions of the manufacturer.
In order to confirm the proper functioning of nucleic acid extraction and reverse transcription steps, as much as 18 µL xTAG bacteriophage MS2 needed to be injected into 182 µL of sample. Following that, complementary DNA (cDNA) synthesis and PCR amplification were conducted according to instructions. The assay was performed in a Bio-Rad DNA Engine (MJ PTC 200) (Bio-Rad, USA) and the mixture was later incubated at 45 °C for 20 minutes in the same machine. Lastly, Luminex xMAP instrument was used for virus detection (Luminex, USA) (18).
Statistical analysis
The data were analyzed with SPSS Statistics 18 version 18.0.0 (SPSS, Inc., 2001, USA). Distribution normality was checked for all variables including age and gender. Due to the unequal group sizes and the data variance, one-way Analysis of variance (ANOVA) with Games-Howell post-hoc test was used to analyze the various age groups of children infected by different viruses. A P value of <0.05 was taken as statistically significant level. The association between categorical data such as age, gender, and ethnic groups with respiratory viruses were analyzed using Fisher’s exact test.
Results
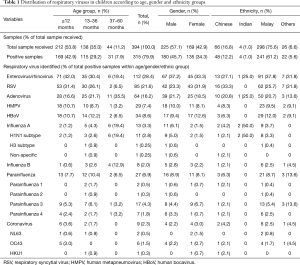
A total of 394 samples were collected in this study. Table 1 outlines the socio-demographic variables of all the sample population. The age distribution was as follows: less than 12 months (212/394, 53.8%), 13–36 months (138/394, 35.0%) and 37–60 months (44/394, 11.2%) (Table 1). The ratio of male:female patients was 1.3. The mean age of the study population was 1.89±1.71 years old, with majority of the patients (53.8%) being less than 1 year old. The viral detection rate was 79.9%, in which 315 out of the total 380 samples were tested positive. As high as 380 viruses were found among 315 (79.9%) positive specimens, Among the viruses detected, there were 15 types of RNA viruses and a single type of DNA virus.
Full table
The prevalence of different viruses detected was also presented in Table 1. The 5 predominant viruses were enterovirus/rhinovirus (n=112, 28.4%), RSV (n=85, 21.6%), adenovirus (n=64, 16.2%), human bocavirus (n=34, 8.6%), and human metapneumovirus (n=29, 7.4%). Among the influenza cases, 13 (3.3%) were influenza A and 8 (2.0%) were influenza B. The 27 typeable parainfluenza viruses can be categorized into 2 cases (0.5%) of parainfluenza 1 virus, 1 case (0.3%) of parainfluenza 2 virus, 17 cases (4.3%) of parainfluenza 3 virus, and 7 cases (1.8%) of parainfluenza 4 virus. A total of 8 coronavirus cases were detected, including coronavirus OC43 (n=5, 1.3%), coronavirus NL63 (n=2, 0.5%), and coronavirus HKU1 (n=1, 0.3%). No coronavirus 229E was detected in the study.
Children aged below 1 year old constituted 42.9% of all the samples received in this study. In this age group, enterovirus/rhinovirus (P<0.05) and RSV (P<0.01) were significantly more common in this age group, accounting for 42.0% and 31.4% of the total cases respectively. With advancing age, the prevalence of enterovirus/rhinovirus and RSV decreased to 19.4% and 6.5% among children aged 4–5 years old. In contrast, influenza viruses increased significantly reduced with age from 3.0% in the age group below 1 year old to 51.7% among the 3–5 years old age groups (P<0.01). Coronavirus was not detected in any children between 3 and 5 years old. The mean ages of infected children ranged from 1.30 to 3.13 years and were significantly different from one-way ANOVA analysis (F=5.401, df=7, P<0.001). Post-hoc test showed that children tested positive for influenza virus were significantly older than children infected with enterovirus/rhinovirus (mean difference =1.44, SE =0.29, P<0.001), RSV (mean difference =1.67, SE =0.29, P<0.001), adenovirus (mean difference =1.15, SE =0.30, P<0.01), human metapneumovirus (mean difference =1.82, SE =0.30, P<0.001), and coronavirus (mean difference =1.97, SE =0.37, P<0.01).
From all the positive samples, 45.7% were male and 34.3% were female patients. Boys were significantly more susceptible to influenza A than female children (P<0.05). However, differences between gender were not statistically significant for the remaining viruses. The predominant ethnicity was Malay (75.6%), followed by Chinese (16.8%), others (6.6%), and Indians (1.0%). There was a significant difference between ethnic groups in all respiratory viruses (P<0.001). Malay population was more likely to be positive for all respiratory viruses.
In Table 2, the different types of viruses detected were shown. About 1 in 5 patients was found to be having multiple viral infections (n=58, 14.7%). In 51 cases, 2 viruses were detected simultaneously. Another 6 patients with enterovirus/rhinovirus were identified to have 2 other viruses, namely RSV, human bocavirus, human metapneumovirus, adenovirus, and coronavirus NL63 (Table 2). Enterovirus/rhinovirus (n=13, 22.4%) was the most frequently detected virus in concurrent with adenovirus. Multiple viral infections were predominantly detected in children less than 1 year old (n=31, 53.4%). No co-infection was detected in young children. One caveat of this study was the lack of clarity as to which viral agent was the causative agent in patients with co-infection. Moreover, the clinical presentation of patients was not assessed and compared to respiratory viral pathogens identified.

Full table
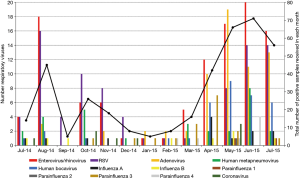
The seasonal distribution of the respiratory viruses from July 2014 to July 2015 is shown in Figure 1. Viral respiratory infections were present throughout the year, with monthly peaks of activity. Enterovirus/rhinovirus, adenovirus, and human bocavirus demonstrated pronounced seasonality, with peak infection rate occurred in mid-year (from April 2015 to July 2015), and lowest activity in early of 2015 (January–March). RSV infections gradually decreased from August 2014 to year-end but subsequently increased from early 2015 to the peak in June–July 2015. The number of other viruses was too small to detect any seasonality.
Discussion
There is limited evidence on epidemiological burden of respiratory viruses in Malaysia in the published literature. This could be attributed to a lack of diagnostic capacity for viral detection (19-21). From the literature, previous studies focused on narrower virus spectrum, including only RSV, adenovirus, influenza virus, and/or parainfluenza virus (13-15). Different geographical areas and detection methods could significantly affect the observed burden of each respiratory virus in different studies.
With modern technology, molecular detection of viruses is becoming more robust and reliable, with better sensitivity and specificity (7,22). The conventional methods such as viral isolation and immunofluorescence staining often underestimated the actual burden of respiratory viruses (15). The use of Luminex xTAG RVP Fast v2 assay in the study allowed the detection of a broader range of respiratory viruses (23). This study revealed that the assay was more sensitive as it has a higher detection rate (78.4%), compared to the viral isolation (7.5%) and direct immunofluorescence staining (22.6%) methods (24).
Previous epidemiological data showed that RSV was the most predominant respiratory virus detected in young children worldwide (5,12,15,25-28). However, our data found that enterovirus/rhinovirus was the commonest virus in our paediatric population, especially among infants (less than 12 months old). This finding was in contrast with studies conducted in other Asian countries whereby RSV was the predominant respiratory virus (5,12,15,25,26). It could be due to more sensitive molecular detection methods being used or the seasonal variations nature of virus distributions. Rhinovirus was neither detected nor included in most of the local studies (15). Our results aligned with a Vietnamese study in which enterovirus/rhinovirus was found to be the most predominant viral agent among young children, followed by RSV (29).
Several emerging respiratory viruses including human bocavirus (8.6%), human metapneumovirus (7.4%), and influenza A virus H1N1 (2.8%) were detected in this study. These affected children may require extra medical care because these 3 viruses had been linked with substantial morbidity in previous studies (30-33). Human coronaviruses, i.e., 229E, HKU1, NL63, and OC43 strains are associated with a range of respiratory infections (34). We detected a total of 8 coronavirus-positive specimens but no coronavirus 229E in this study. Coronavirus 229E differs clinically from other respiratory viruses (34) and it is often associated with immunocompromised individuals (35,36).
This study included young children between age below 5 years old. The burden of viral respiratory infections is higher in infants (less than 12 months) as a result of an immature immune system and waning maternal antibodies after 6 months of life to prevent viral infections. With age, children would acquire immunity from repeated infections, thus making them less susceptible to viral infections.
Similar to a previous study, the coronavirus-positive specimens were detected in children below the age of 3 (37). This could be attributed to the fact that maternal antibodies provide protection against coronavirus NL63 and 229E only during the first 3 months of life (38). Subsequently, children are susceptible to coronavirus NL63 and 229E infections until they are able to develop antibodies between the age of 2.5 to 3.5 years old (38). In concordance with a previous study (15), we also observed that boys were more vulnerable (45.7%) to ARTIs than girls.
Geographically, Malaysia is located in the central region of South East Asia. It borders Singapore on the South, Thailand on the north, and Indonesia to the South and West. High temperatures and humidity are usually recorded throughout the year. Based on our findings, the highest burden of viral infection (18.0%) was recorded in June. As for different viruses, the enterovirus/rhinovirus and RSV caused endemic infections as they were detected throughout the study period. Previous local studies showed that RSV peaked during August 2014, June 2015, and July 2015 in Kuala Lumpur (12,15). Studies from Thailand and Lombok, Indonesia revealed a peak in RSV infection from March to August (39,40). Some studies linked RSV infections with the rainy season (15,41) but the association with humidity was less clear (15,41,42). In contrast to previous studies (12,43), we observed no seasonality for influenza and parainfluenza viruses, probably due to the small number of infections by these 2 viruses in our study.
There are 2 main limitations in this study. The first caveat was the lack of clarity as to which viral agent was the causative agent in patients with co-infection. Moreover, the clinical presentation of patients was not assessed and compared to respiratory viral pathogens identified. One of the limitations includes the unclear viruses that cause the clinical manifestations of the ARTIs owe to a lack of patients’ clinical history.
Conclusions
In summary, this study provided important molecular surveillance of ARTIs burden and seasonality between 2014 and 2015 among young children in Malaysia. The variety of respiratory viral pathogens detected in this study highlighted the need for a monitoring database of ARTIs and its aetiological pathogen among young children. This data would provide useful guidance for healthcare professionals to implement the necessary prevention and management strategies for young children with ARTIs in Malaysia.
Acknowledgments
Funding: This study was supported by High Impact Research MoE Grant UM.C/625/1/ HIR/MOHE/MED/31 (No. H-20001-00-E000070).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Medical Ethics Committee of University Malaya Medical Centre (reference number: 20148-438) and written informed consent was obtained from all patients.
References
- Williams BG, Gouws E, Boschi-Pinto C, et al. Estimates of world-wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis 2002;2:25-32. [Crossref] [PubMed]
- Regamey N, Kaiser L, Roiha HL, et al. Viral etiology of acute respiratory infections with cough in infancy: a community-based birth cohort study. Pediatr Infect Dis J 2008;27:100-5. [PubMed]
- Bush A, Thomson AH. Acute bronchiolitis. BMJ 2007;335:1037-41. [Crossref] [PubMed]
- Black RE, Cousens S, Johnson HL, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 2010;375:1969-87. [Crossref] [PubMed]
- Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 2010;23:74-98. [Crossref] [PubMed]
- Meerhoff TJ, Mosnier A, Schellevis F, et al. Progress in the surveillance of respiratory syncytial virus (RSV) in Europe: 2001-2008. Euro Surveill 2009;14. [PubMed]
- Iwane MK, Edwards KM, Szilagyi PG, et al. Population-based surveillance for hospitalizations associated with respiratory syncytial virus, influenza virus, and parainfluenza viruses among young children. Pediatrics 2004;113:1758-64. [Crossref] [PubMed]
- Song JR, Jin Y, Xie ZP, et al. Novel human bocavirus in children with acute respiratory tract infection. Emerg Infect Dis 2010;16:324-7. [Crossref] [PubMed]
- Monto AS, Koopman JS, Bryan ER. The Tecumseh Study of Illness. XIV. Occurrence of respiratory viruses, 1976-1981. Am J Epidemiol 1986;124:359-67. [Crossref] [PubMed]
- Jin Y, Zhang RF, Xie ZP, et al. Prevalence of adenovirus in children with acute respiratory tract infection in Lanzhou, China. Virol J 2013;10:271. [Crossref] [PubMed]
- Singleton RJ, Holman RC, Folkema AM, et al. Trends in lower respiratory tract infection hospitalizations among American Indian/Alaska Native children and the general US child population. J Pediatr 2012;161:296-302.e2. [Crossref] [PubMed]
- Zamberi S, Zulkifli I, Ilina I. Respiratory viruses detected in hospitalised paediatric patients with respiratory infections. Med J Malaysia 2003;58:681-7. [PubMed]
- Chan PW, Chew FT, Tan TN, et al. Seasonal variation in respiratory syncytial virus chest infection in the tropics. Pediatr Pulmonol 2002;34:47-51. [Crossref] [PubMed]
- Saat Z, Abdul Rashid TR, Yusof MA, et al. Seasonal influenza virus strains circulating in Malaysia from 2005 to 2009. Southeast Asian J Trop Med Public Health 2010;41:1368-73. [PubMed]
- Khor CS, Sam IC, Hooi PS, et al. Epidemiology and seasonality of respiratory viral infections in hospitalized children in Kuala Lumpur, Malaysia: a retrospective study of 27 years. BMC Pediatr 2012;12:32. [Crossref] [PubMed]
- Kuan CS, Sim HP, Lee LM, et al. Comparison of Anyplex II RV16 assay with conventional methods for detection of respiratory viruses. Trop Biomed 2016;33:311-9.
- Henrickson KJ, Kuhn SM, Savatski LL, et al. Recovery of human parainfluenza virus types one and two. J Virol Methods 1994;46:189-205. [Crossref] [PubMed]
- Bortolin S, Black M, Modi H, et al. Analytical validation of the tag-it high-throughput microsphere-based universal array genotyping platform: application to the multiplex detection of a panel of thrombophilia-associated single-nucleotide polymorphisms. Clin Chem 2004;50:2028-36. [Crossref] [PubMed]
- LaSala PR, Bufton KK, Ismail N, et al. Prospective comparison of R-mix shell vial system with direct antigen tests and conventional cell culture for respiratory virus detection. J Clin Virol 2007;38:210-6. [Crossref] [PubMed]
- Liolios L, Jenney A, Spelman D, et al. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol 2001;39:2779-83. [Crossref] [PubMed]
- Tantivanich S, Suphanaranonda K, Balachanda K, et al. Detection of respiratory syncytial virus from clinical specimens: comparison between reverse transcription polymerase chain reaction and tissue culture. Southeast Asian J Trop Med Public Health 1995;26:684-8. [PubMed]
- Thomazelli LM, Vieira S, Leal AL, et al. Surveillance of eight respiratory viruses in clinical samples of pediatric patients in southeast Brazil. J Pediatr (Rio J) 2007;83:422-8. [Crossref] [PubMed]
- Merante F, Yaghoubian S, Janeczko R. Principles of the xTAG respiratory viral panel assay (RVP Assay). J Clin Virol 2007;40 Suppl 1:S31-5. [Crossref] [PubMed]
- Kuan CS, Yew SM, Hooi PS, et al. Detection of Respiratory Viruses from ARTI Patients by xTAG RVP Fast v2 Assay and Conventional Methods. Malays J Med Sci 2017;24:33-43. [Crossref] [PubMed]
- Choi EH, Lee HJ, Kim SJ, et al. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000-2005. Clin Infect Dis 2006;43:585-92. [Crossref] [PubMed]
- Yeolekar LR, Damle RG, Kamat AN, et al. Respiratory viruses in acute respiratory tract infections in Western India. Indian J Pediatr 2008;75:341-5. [Crossref] [PubMed]
- Kwofie TB, Anane YA, Nkrumah B, et al. Respiratory viruses in children hospitalized for acute lower respiratory tract infection in Ghana. Virol J 2012;9:78. [Crossref] [PubMed]
- Shafik CF, Mohareb EW, Yassin AS, et al. Viral etiologies of lower respiratory tract infections among Egyptian children under five years of age. BMC Infect Dis 2012;12:350. [Crossref] [PubMed]
- Yoshida LM, Suzuki M, Yamamoto T, et al. Viral pathogens associated with acute respiratory infections in central vietnamese children. Pediatr Infect Dis J 2010;29:75-7. [Crossref] [PubMed]
- Loo LH, Tan BH, Ng LM, et al. Human metapneumovirus in children, Singapore. Emerg Infect Dis 2007;13:1396-8. [Crossref] [PubMed]
- Samransamruajkit R, Thanasugarn W, Prapphal N, et al. Human metapneumovirus in infants and young children in Thailand with lower respiratory tract infections; molecular characteristics and clinical presentations. J Infect 2006;52:254-63. [Crossref] [PubMed]
- Tan BH, Lim EA, Seah SG, et al. The incidence of human bocavirus infection among children admitted to hospital in Singapore. J Med Virol 2009;81:82-9. [Crossref] [PubMed]
- Bearden A, Friedrich TC, Goldberg TL, et al. An outbreak of the 2009 influenza a (H1N1) virus in a children's hospital. Influenza Other Respir Viruses 2012;6:374-9. [Crossref] [PubMed]
- Gaunt ER, Hardie A, Claas EC, et al. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol 2010;48:2940-7. [Crossref] [PubMed]
- Gerna G, Campanini G, Rovida F, et al. Genetic variability of human coronavirus OC43-, 229E-, and NL63-like strains and their association with lower respiratory tract infections of hospitalized infants and immunocompromised patients. J Med Virol 2006;78:938-49. [Crossref] [PubMed]
- Pene F, Merlat A, Vabret A, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis 2003;37:929-32. [Crossref] [PubMed]
- Talbot HK, Crowe JE Jr, Edwards KM, et al. Coronavirus infection and hospitalizations for acute respiratory illness in young children. J Med Virol 2009;81:853-6. [Crossref] [PubMed]
- Dijkman R, Jebbink MF, El Idrissi NB, et al. Human coronavirus NL63 and 229E seroconversion in children. J Clin Microbiol 2008;46:2368-73. [Crossref] [PubMed]
- Chew FT, Doraisingham S, Ling AE, et al. Seasonal trends of viral respiratory tract infections in the tropics. Epidemiol Infect 1998;121:121-8. [Crossref] [PubMed]
- Djelantik IG, Gessner BD, Soewignjo S, et al. Incidence and clinical features of hospitalization because of respiratory syncytial virus lower respiratory illness among children less than two years of age in a rural Asian setting. Pediatr Infect Dis J 2003;22:150-7. [Crossref] [PubMed]
- Omer SB, Sutanto A, Sarwo H, et al. Climatic, temporal, and geographic characteristics of respiratory syncytial virus disease in a tropical island population. Epidemiol Infect 2008;136:1319-27. [Crossref] [PubMed]
- Loh TP, Lai FY, Tan ES, et al. Correlations between clinical illness, respiratory virus infections and climate factors in a tropical paediatric population. Epidemiol Infect 2011;139:1884-94. [Crossref] [PubMed]
- Suwanjutha S, Chantarojanasiri T, Watthana-kasetr S, et al. A study of nonbacterial agents of acute lower respiratory tract infection in Thai children. Rev Infect Dis 1990;12 Suppl 8:S923-8. [Crossref] [PubMed]


