
Identification of key genes and pathways in calcific aortic valve disease by bioinformatics analysis
Introduction
Calcific aortic valve disease (CAVD) is the most common type of valvular heart disease in the elderly (1). Calcific aortic valve stenosis occurred in 2% of people above 65 years old; while the incidence of aortic valve sclerosis, the early stage of CAVD without hemodynamic consequence, was as high as 26% (2). Even without hemodynamically significant obstruction of left ventricular outflow, CAVD still can be an independent risk factor for cardiovascular mortality, all-cause mortality and myocardial infarction (1,3). Currently, the only effective therapeutic option to improve clinical outcomes in CAVD patients is early surgery (4).
Histological studies suggested that the early lesion of CAVD was an active inflammatory process shared some similarities with atherosclerosis, such as lipid deposition, macrophage and T-cell infiltration, and basement membrane disruption (5). While the end-stage of CAVD usually involved heterotopic ossification consisting of mature lamellar bone formation and active bone remodeling (6). Several serum biomarkers such as lipoprotein(a) (2,7), LDL-C (2), and serum phosphate (8) had been found to be associated with the prevalence of CAVD. Various molecular, cellular, and biomechanical mechanisms had been found involved in the progression of CAVD, such as bicuspid aortic valve, osteogenic mechanisms, endocrine mechanisms, dysregulated mineral metabolism, osteoclast deficiency, and developmental signaling (9). Growing evidences suggested that the progression of CAVD was a actively regulated process, which might be suspended or reversed via potential pharmacologic intervention (10). However, thus far, there have been no identified pharmacologic interventions or presumably underlying pathways that affect the CAVD progression process. Besides, the key genes and signaling pathways underlying the progression of CAVD remain poorly understood.
Gene expression microarrays have been widely used to study gene expression profiles in many human diseases, which provide an effective way for studying disease-associated genes and discovering novel therapeutic targets and biomarkers (11). Recently, several studies had compared the mRNA expression profile of CAVD (12,13). However, the results were inconsistent due to small sample size, different technological platforms, different data processing and sample heterogeneity. The disadvantage of these single studies can be overcome by integrated bioinformatics analysis, which combines different microarray dataset to obtain a more steady result (14). MetaDE package developed by Wang et al. contains more than 12 popular genomic meta-analysis methods, which is an effective integrative analysis solution (15).
In the present study, we applied integrated bioinformatics methods to identify key genes and pathways in CAVD. Two microarray profile datasets [GSE51472 (12) and GSE83453 (13)] were downloaded from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were screened using the MetaDE package in R software. Gene ontology (GO) and pathway enrichment analysis of DEGs were then performed. A protein-protein interaction (PPI) network was established, and key genes and pathways associated with CAVD were analyzed.
Methods
Microarray data
The present study was a bioinformatics study based on publicly accessible microarray datasets. Thus, an ethics approval was not required. The gene expression profiles of GSE51472 (12) and GSE83453 (13) were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The platform for GSE51472 is GPL570, Affymetrix Human Genome U133 Plus 2.0 Array, which includes 5 normal aortic valve samples, 5 fibrotic aortic valve samples and 5 calcified aortic valve samples. The control group contained aortic valves from patients underwent surgery due to ascending aortic aneurysm or dissection, while the valves were macroscopically smooth, pliable and opalescent, and without visible calcifications. The fibrotic group contained valves that were macroscopically thickened and stiff, while the patients had no significant transvalvular pressure gradient. The calcified group contained aortic valves from patients with severe aortic valve stenosis, and the valves were presented with varying degrees of calcification. Only the normal and calcified aortic valve samples were used for our bioinformatics analysis. The platform for GSE83453 is GPL10558, Illumina HumanHT-12 V4.0 expression beadchip, which consists of 8 normal tricuspid aortic valve samples, 10 calcified bicuspid aortic valve samples and 9 calcified tricuspid aortic valve samples. The calcified bicuspid and tricuspid aortic valve samples were merged to build the calcified aortic valve group in our bioinformatics analysis. The dataset information is shown in Table 1. The data were normalized and log2 transformed.

Full table
Data preprocessing
The probe IDs was converted into international standard names for genes (gene symbol). For multiple probes that mapped to a single gene, interquartile range (IQR) of each probe was calculated with MetaDE.match package in R software, and the probe with largest IQR was selected to represent the expression level of the gene. Next, MetaDE.merge package was used to merge the gene expression profiles to obtain the commonly profiles genes between the two datasets. Finally, either 30% un-expressed genes (small mean intensity) or 30% un-informative genes (small standard deviation) were filtered out by MetaDE.filtering package, and the remaining 7,946 genes were remained for further analysis.
Integration of microarray data
MetaDE package was used to perform the integrate analysis (15). Moderate t test was used to determine the gene expression difference in each dataset, and the maximum P value method (maxP) was used to combine the P values from moderate t test. An adjusted P value of <0.0001, based on the false discovery rate (FDR) using Benjamini-Hochberg method was used to select DEGs.
GO and pathway enrichment analysis of DEGs
GO annotations were performed using DAVID database (https://david.ncifcrf.gov/) (16). KEGG (17) pathway enrichment analysis was performed using the KOBAS online analysis database (http://kobas.cbi.pku.edu.cn/). Up- and down-regulated DEGs were subjected to GO and pathway enrichment analysis separately. P value <0.05 was used as threshold value.
PPI network analysis
The STRING database (http://string-db.org/) (18) was used to identify the interactions between proteins encoded by DEGs based on experimental data, databases, text mining, and predictive bioinformatics data. A combined score of >0.4 was set as threshold value. PPI networks were constructed with Cytoscape software (19). The genes with higher degrees of interaction were considered as hub genes, and pathway enrichment analysis was performed for these hub genes. The plug-in Molecular Complex Detection (MCODE) was used to screen the modules of PPI network in Cytoscape software. MCODE score >3 and number of nodes >4 were considered as threshold value. Pathway enrichment analysis were performed for genes in the modules.
Results
Identification of DEGs
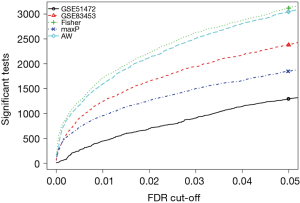
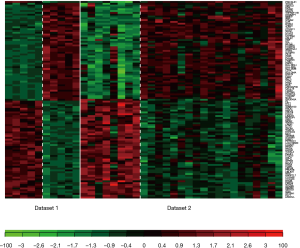
Three method were used to detect common DEGs between the two microarray datasets: the maximum P value method (maxP), the Fisher’s method (Fisher), and the adaptive weight method (AW). The detection competency curves (Figure 1) suggested all the three methods were useful to detect common DEGs between the two datasets. The maxP method was chosen in this study. Under the threshold of adjusted P value of <0.0001, 107 DEGs were identified in CAVD. Among the 107 DEGs, 53 were up-regulated genes, and 54 were down-regulated genes (Table 2). Heatmap showed the expression pattern of these DEGs (Figure 2), suggesting the consistency of the 107 DEGs in each dataset.

Full table
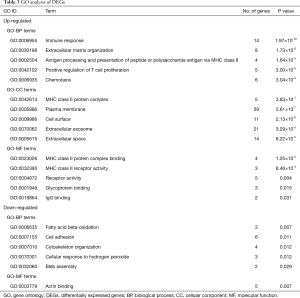
GO analysis
GO terms were divided into three functional groups: biological processes (BP), cellular component (CC), and molecular function (MF). The top 5 enriched GO terms in each category were listed in Table 3. For BP, the up-regulated DEGs were mainly enriched in GO terms of immune response, extracellular matrix organization, antigen processing and presentation via MHC class II, positive regulation of T cell proliferation, and chemotaxis; while the down-regulated DEGs were mainly enriched in GO terms of fatty acid beta-oxidation, cell adhesion, cytoskeleton organization, cellular response to hydrogen peroxide. For CC, the up-regulated DEGs were mainly enriched in GO terms of MHC class II protein complex, plasma membrane, cell surface, extracellular exosome, and extracellular space; while no GO-CC term was enriched from down-regulated DEGs. For MF, the up-regulated DEGs were mainly enriched in GO terms of MHC class II binding and receptor activity.
Full table
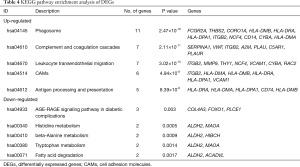
Pathway enrichment analysis
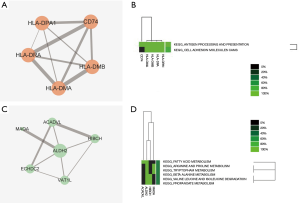
The most significantly enriched pathways of DEGs analyzed by KEGG analysis were listed in Table 4. The up-regulated DEGs were enriched in phagosome, complement and coagulation cascades, leukocyte transendothelial migration, cell adhesion molecules (CAMs), and antigen processing and presentation. It was noteworthy that four HLA molecules (HLA-DRA, HLA-DMA, HLA-DPA1, HLA-DMB) presented in three pathways associated with immune response, while ITGB2 and VCAM1 presented in two pathways associated with cell adhesion and migration. The down-regulated DEGs were enriched in AGE-RAGE signaling pathway, metabolism of amino acid (histidine, beta-Alanine, tryptophan) and fatty acid. ALDH2 presented in metabolism of amino acids and fatty acid.
Full table
PPI network analysis
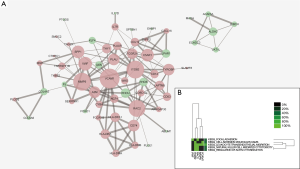
A total of 61 nodes and 143 protein pairs were obtained with a combined score of >0.4 based on the STRING database (Figure 3A). The top 5 hub genes a degree of interaction ≥10 were VCAM1 (degree =16), MMP9 (degree =14), ITGB2 (degree =14), RAC2 (degree =13), and vWF (degree =10). Pathway enrichment analysis suggested that these hub genes mainly involved in leukocyte transendothelial migration and cell adhesion (Figure 3B). MCODE detected a significant protein cluster containing HLA-DPA1, HLA-DRA, HLA-DMA, HLA-DMB, and CD74 (Figure 4A). Pathway enrichment analysis showed that these genes mainly involved in antigen processing and presentation and CAMs (Figure 4B). We also conducted pathway enrichment analysis to the independently down-regulated protein cluster containing ALDH2, HIBCH, ACADVL, ECHDC2, VAT1L, and MAOA (Figure 4C), which suggested that these genes mainly involved in metabolism of fatty acid and various amino acids (Figure 4D).
Discussion
In the present study, a total of 107 DEGs (53 up and 54 down-regulated genes) were identified in CAVD by integrated analysis of microarray datasets (GSE51472 and GSE83453). Further bioinformatics analysis suggested that VCAM1, MMP9, ITGB2, RAC2, and vWF had a high degree of interaction in the PPI network and were key genes in CAVD. These key genes were significantly enriched in pathway of leukocyte transendothelial migration. The gene cluster containing HLA-DPA1, HLA-DRA, HLA-DMA, HLA-DMB, and CD74 detected by MCODE also suggested the role of immune response in CAVD. Moreover, the down-regulated gene ALDH2 might play a protective role in CAVD.
Inflammation is involved in the development and progression of CAVD (20), and calcification of the aortic valve was considered an inflammation-dependent process (21). T lymphocytes (22) and macrophages (5) had been identified in early aortic valve lesions. In the present study, 4 hub genes (VCAM1, MMP9, ITGB2, and RAC2) involved in leukocyte transendothelial migration, which was the first step of the immune cells infiltration. Aortic valve endothelial cells cover the surface of the aortic valve, and provide a protective function on aortic valve (23,24). High expression of VCAM1 had been found in the endothelium of calcified human aortic valves (24). VCAM1 on the endothelial cells are involved in the firm adhesion of leukocytes (25), and it also activates intracellular calcium release and NADPH oxidase Nox2, which further promotes the migration of leukocytes (26). Increase in MMP-9 had been found in aortic stenosis (27). MMP9 is synthesized by macrophages, which regulates the activation of T lymphocytes (28). MMP9 also can regulate leukocyte migration by processing chemokines and cytokines (29). ITGB2 normally expressed only in leukocytes, which promote leukocyte adhesion to endothelium (30). RAC2 is a member of GTPase signaling, which had been proven involved in regulation of leukocyte transendothelial migration (31). However, the role of ITGB2 and RAC2 in CAVD has not been illuminated yet. HLA-DPA1, HLA-DRA, HLA-DMA, HLA-DMB, and CD74 are MHC class II molecules, which mainly expressed in antigen presenting cells such as macrophages and dendritic cells (32). It had been previously reported that the proportion of circulating CD8+ T cell expressing HLA-DR was elevated during CAVD, which suggested that besides leukocytes infiltration of the aortic valve, a systemic adaptive immunity also play a role during CAVD (33).
vWF plays a major role in platelet adhesion and hemostasis (34). An in vitro study conducted by Balaoing et al. showed that vWF stimulated by histamine from porcine aortic valve endothelial cells significantly increased valvular interstitial cell calcification in vitro, which suggested a possible role of vWF in progression of CAVD (35).
ALDH2 is a detoxifying enzyme for the removal of toxic acetaldehyde such as malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE) (36). More than 40% of the East Asians population carries a common ALDH2*2 mutant allele (Glu504Lys), which results in reduction of the enzymatic activity (37). Recently, various studies had highlighted the relationship between ALDH2 and cardiovascular disease. Epidemiological studies suggested that the inactive East-Asian variant of ALDH2 (ALDH2*2) increased the risk of coronary artery disease (38). Animal study suggested that Alda-1, an activator of ALDH2, inhibited atherosclerosis in apolipoprotein E-knockout mice (39). In vitro study also suggested that activation of ALDH2 significantly attenuated the oxygenized LDL induced endoplasmic reticulum stress and apoptosis in smooth muscle cells (40). The present study suggested that the expression of ALDH2 was down-regulated in CAVD, however, the role of ALDH2 in CAVD had not been reported yet. It has been demonstrated that dysregulation of antioxidant mechanisms led to increased oxidative stress, which contributed to the progression of CAVD (41). We speculated that ALDH2-mediated detoxification effect reduced the oxidative stress in aortic valve, which might play a protective role in CAVD.
There are several limitations of the present study. The sample sizes of the microarray datasets used in the integrated analysis were relatively small. Besides, due to the very small sample sizes, subgroup analysis was not able to be performed to distinguish the possible different mechanisms involved between bicuspid and tricuspid valves. Although the integrated bioinformatics approach obtained a steadier result than single microarray, further studies with larger sample sizes covering wider range of races are still needed.
In conclusion, the present study identified VCAM1, MMP9, ITGB2, RAC2, vWF and ALDH2 as key genes in the progression of CAVD by integrated analysis of microarray datasets. Further bioinformatics analysis suggested that immune cells infiltration might play a key role in the progression of CAVD, while ALDH2-mediated detoxification effect might play a protective role in CAVD. The study provides a set of useful targets for further investigation into the molecular mechanisms, biomarkers and treatment targets of CAVD. Further molecular biological experiments are needed to confirm the function of the identified genes associated with CAVD.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant No. 81670350) and Natural Science Foundation of Zhejiang Province, China (Grant No. LQ20H020003).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Otto CM, Lind BK, Kitzman DW, et al. Association of aortic-valve sclerosis with cardiovascular mortality and morbidity in the elderly. N Engl J Med 1999;341:142-7. [Crossref] [PubMed]
- Stewart BF, Siscovick D, Lind BK, et al. Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol 1997;29:630-4. [Crossref] [PubMed]
- Blaha MJ, Budoff MJ, Rivera JJ, et al. Relation of aortic valve calcium detected by cardiac computed tomography to all-cause mortality. Am J Cardiol 2010;106:1787-91. [Crossref] [PubMed]
- Kang DH, Park SJ, Rim JH, et al. Early surgery versus conventional treatment in asymptomatic very severe aortic stenosis. Circulation 2010;121:1502-9. [Crossref] [PubMed]
- Otto CM, Kuusisto J, Reichenbach DD, et al. Characterization of the early lesion of 'degenerative' valvular aortic stenosis. Histological and immunohistochemical studies. Circulation 1994;90:844-53. [Crossref] [PubMed]
- Mohler ER 3rd, Gannon F, Reynolds C, et al. Bone formation and inflammation in cardiac valves. Circulation 2001;103:1522-8. [Crossref] [PubMed]
- Thanassoulis G, Campbell CY, Owens DS, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med 2013;368:503-12. [Crossref] [PubMed]
- Linefsky JP, O'Brien KD, Sachs M, et al. Serum phosphate is associated with aortic valve calcification in the Multi-ethnic Study of Atherosclerosis (MESA). Atherosclerosis 2014;233:331-7. [Crossref] [PubMed]
- Yutzey KE, Demer LL, Body SC, et al. Calcific aortic valve disease: a consensus summary from the Alliance of Investigators on Calcific Aortic Valve Disease. Arterioscler Thromb Vasc Biol 2014;34:2387-93. [Crossref] [PubMed]
- Otto CM. Calcific aortic stenosis--time to look more closely at the valve. N Engl J Med 2008;359:1395-8. [Crossref] [PubMed]
- Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet 2013;14:89-99. [Crossref] [PubMed]
- Ohukainen P, Syvaranta S, Napankangas J, et al. MicroRNA-125b and chemokine CCL4 expression are associated with calcific aortic valve disease. Ann Med 2015;47:423-9. [Crossref] [PubMed]
- Guauque-Olarte S, Droit A, Tremblay-Marchand J, et al. RNA expression profile of calcified bicuspid, tricuspid, and normal human aortic valves by RNA sequencing. Physiol Genomics 2016;48:749-61. [Crossref] [PubMed]
- Ramasamy A, Mondry A, Holmes CC, et al. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med 2008;5:e184. [Crossref] [PubMed]
- Wang X, Kang DD, Shen K, et al. An R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics 2012;28:2534-6. [Crossref] [PubMed]
- Sherman BT. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 2007;8:426. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Coté N, Mahmut A, Bosse Y, et al. Inflammation is associated with the remodeling of calcific aortic valve disease. Inflammation 2013;36:573-81. [Crossref] [PubMed]
- Dweck MR, Khaw HJ, Sng GK, et al. Aortic stenosis, atherosclerosis, and skeletal bone: is there a common link with calcification and inflammation? Eur Heart J 2013;34:1567-74. [Crossref] [PubMed]
- Olsson M, Dalsgaard CJ, Haegerstrand A, et al. Accumulation of T lymphocytes and expression of interleukin-2 receptors in nonrheumatic stenotic aortic valves. J Am Coll Cardiol 1994;23:1162-70. [Crossref] [PubMed]
- Richards J, El-Hamamsy I, Chen S, et al. Side-specific endothelial-dependent regulation of aortic valve calcification: interplay of hemodynamics and nitric oxide signaling. Am J Pathol 2013;182:1922-31. [Crossref] [PubMed]
- Farrar EJ, Huntley GD, Butcher J. Endothelial-derived oxidative stress drives myofibroblastic activation and calcification of the aortic valve. PLoS One 2015;10:e0123257. [Crossref] [PubMed]
- Barreiro O, Yanez-Mo M, Serrador JM, et al. Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J Cell Biol 2002;157:1233-45. [Crossref] [PubMed]
- Muller WA. Mechanisms of leukocyte transendothelial migration. Annu Rev Pathol 2011;6:323-44. [Crossref] [PubMed]
- Fondard O, Detaint D, Iung B, et al. Extracellular matrix remodelling in human aortic valve disease: the role of matrix metalloproteinases and their tissue inhibitors. Eur Heart J 2005;26:1333-41. [Crossref] [PubMed]
- Benson HL, Mobashery S, Chang M, et al. Endogenous matrix metalloproteinases 2 and 9 regulate activation of CD4+ and CD8+ T cells. Am J Respir Cell Mol Biol 2011;44:700-8. [Crossref] [PubMed]
- Van Lint P, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol 2007;82:1375-81. [Crossref] [PubMed]
- Tan SM. The leucocyte beta2 (CD18) integrins: the structure, functional regulation and signalling properties. Biosci Rep 2012;32:241-69. [Crossref] [PubMed]
- Wittchen ES, van Buul JD, Burridge K, et al. Trading spaces: Rap, Rac, and Rho as architects of transendothelial migration. Curr Opin Hematol 2005;12:14-21. [Crossref] [PubMed]
- Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell 2002;109 Suppl:S21-33. [Crossref] [PubMed]
- Winchester R, Wiesendanger M, O'Brien W, et al. Circulating activated and effector memory T cells are associated with calcification and clonal expansions in bicuspid and tricuspid valves of calcific aortic stenosis. J Immunol 2011;187:1006-14. [Crossref] [PubMed]
- Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem 1998;67:395-424. [Crossref] [PubMed]
- Balaoing LR, Post AD, Liu H, et al. Age-related changes in aortic valve hemostatic protein regulation. Arterioscler Thromb Vasc Biol 2014;34:72-80. [Crossref] [PubMed]
- Bosron WF, Ehrig T, Li TK. Genetic factors in alcohol metabolism and alcoholism. Semin Liver Dis 1993;13:126-35. [Crossref] [PubMed]
- Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci U S A 1984;81:258-61. [Crossref] [PubMed]
- Zhang LL, Wang YQ, Fu B, et al. Aldehyde dehydrogenase 2 (ALDH2) polymorphism gene and coronary artery disease risk: a meta-analysis. Genet Mol Res 2015;14:18503-14. [Crossref] [PubMed]
- Stachowicz A, Olszanecki R, Suski M, et al. Mitochondrial aldehyde dehydrogenase activation by Alda-1 inhibits atherosclerosis and attenuates hepatic steatosis in apolipoprotein E-knockout mice. J Am Heart Assoc 2014;3:e001329. [Crossref] [PubMed]
- Yang MY, Wang YB, Han B, et al. Activation of aldehyde dehydrogenase 2 slows down the progression of atherosclerosis via attenuation of ER stress and apoptosis in smooth muscle cells. Acta Pharmacol Sin 2018;39:48-58. [Crossref] [PubMed]
- Miller JD, Chu Y, Brooks RM, et al. Dysregulation of antioxidant mechanisms contributes to increased oxidative stress in calcific aortic valvular stenosis in humans. J Am Coll Cardiol 2008;52:843-50. [Crossref] [PubMed]


