Prognostic role of dysregulated circRNAs in patients with non-small cell lung cancer: a meta-analysis
Introduction
Lung cancer is the most frequent diagnosed cancer and the leading cause of cancer death worldwide, with nearly 2.1 million new cases and 1.8 million deaths in 2018 (1). Lung cancer is composed of small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) according to histopathological type, among which NSCLC accounts for over 85% of all cases (2). Much progress has been achieved in the diagnosis and treatment of NSCLC in recent years, but the 5-year survival rate of NSCLC patients remains unsatisfied, which merely varies from 4–17% depending on the stage and regional differences (3).
Circular RNAs (circRNAs), a novel class of endogenous non-coding RNAs (ncRNAs), were characterized by their abundance, highly conservation, and stable covalently closed continuous loop without free 5’ cap and 3’ tail (4-7). CircRNAs were defined as the by-products of splicing mistakes without biological functions in the past several decades (8). More and more circRNAs have been detected in different cancer tissues with the widely application of high-throughput sequencing technology and bioinformatic approaches (9). Recent studies demonstrated that circRNAs could play a role in a series of biological processes such as microRNA sponging, RBP sponging and mRNA regulation which make effect to carcinogenesis (7,10-12). However, these studies determining the prognostic roles of circRNAs in NSCLC patients were limited by their small sample size. This meta-analysis aimed to evaluate the prognostic value of circRNAs in NSCLC patients.
Methods
Search strategy
This meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (13). Two authors independently searched in PubMed, Web of Science, Embase, and Ovid MEDLINE(R) up to May 20, 2019 using the following search term: (“lung squamous cell carcinoma” OR “lung adenocarcinoma” OR “large cell lung cancer” OR “non-small cell lung cancer” OR NSCLC) AND (“circular RNAs” OR “circular RNA” OR circRNAs OR circRNA) AND (prognosis OR prognostic OR survival).
Inclusion and exclusion criteria
The inclusion criteria were as follows: (I) studies detected circRNAs expression in NSCLC patients; (II) studies investigated the association between circRNAs expression and NSCLC patients’ prognosis; and (III) studies presented hazard ratio (HR) and corresponding 95% confidence interval (CI) or Kaplan-Meier (K-M) curve.
The exclusion criteria were as follows: (I) duplicate publications; (II) reviews, case reports, or letters; (III) studies no published in English; (IV) studies which sample size <35; and (V) studies without available data.
Data extraction
Two authors independently extracted the data from all eligible studies. The following information were extracted from the included studies: first author, publication year, country, sample size, sample types, circRNAs studied, detection methods, circRNAs expression level, cut-off value, number of patients in low expression groups, number of patients in high expression groups, outcome, follow-up period, multivariate analysis, and HR and 95% CI for overall survival (OS). Engauge Digitizer 4.1 software and Tierney’s protocol (14) were applied to extract the HR and 95% CI indirectly from K-M curves if the studies did not present complete survival data.
Quality assessment
Two authors assessed the quality of the included studies independently using Newcastle-Ottawa Scale (NOS) (15). Total scores of NOS were ranged from 0 to 9, and studies with scores greater than 6 were considered as high quality studies.
Statistical analysis
Statistical analyses were analyzed using STATA 12.0 software. Heterogeneity test was performed by I-squared statistics. If I-squared value >50% and P value <0.05, there was significant heterogeneity among the included studies, and a random effects model was applied to estimate the pooled results; otherwise, a fixed effects model was performed. Pooled HR and corresponding 95% CI were calculated to assess the prognostic value of circRNAs in patients with NSCLC. Sensitivity analysis was performed to test the stability of pooled results. Publication bias was estimated using Begg’s funnel plot. Statistical significance was defined as P value <0.05.
Results
Search strategy results
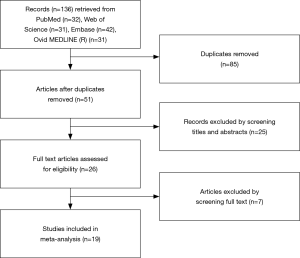
A total of 136 articles were identified from PubMed, Web of Science, Embase, and Ovid MEDLINE (R). After removing 85 duplicate publications, 51 studies were included for further screening. After screening the titles, abstracts, and full texts, 19 eligible studies (16-34) were finally included in current meta-analysis based on the inclusion and exclusion criteria (Figure 1).
Flowchart showing the process of literature search and selection.
Main characteristics and quality assessment of eligible studies
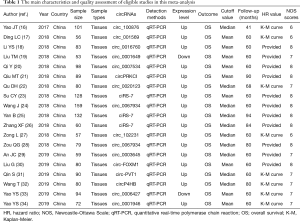
In total, nineteen studies (16-34) comprised of 16 circRNAs were included in this meta-analysis. The main characteristics and quality assessment of the included studies (16-34) were summarized in Table 1. Publication year of all included studies (16-34) ranged from 2017 to 2019. The expression of circRNAs in all included studies (16-34) was detected by quantitative real-time polymerase chain reaction (qRT-PCR). Follow-up period ranged from 41 to 94 months. Eleven studies (18-21,23-26,28-30) directly provided HRs and 95% CI; moreover, we calculated the HRs and 95% CI for the other 8 studies (16,17,22,27,31-34). Furthermore, we assessed the quality of each included studies and the scores ranged from 6–8, which suggests the included studies are in a high quality.
Full table
Prognostic value of circRNAs in patients with NSCLC
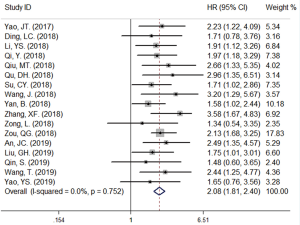
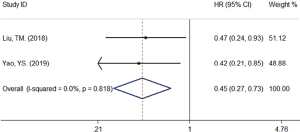
A total of 19 studies (16-34) comprising 1,650 patients with NSCLC were included in the meta-analysis. There were 17 studies (16-18,20-32,34) indicating the association between the up-regulation of circRNAs and the poor prognosis of NSCLC patients. Since there was no significant heterogeneity among the above 17 studies (16-18,20-32,34) (I-squared value =0.0% & P value =0.752), we performed a fixed effects model to assess the pooled HR and corresponding 95% CI. As shown in Figure 2, the up-regulation of circRNAs was significantly associated with the poor prognosis of NSCLC patients (HR =2.08, 95% CI: 1.81–2.40). In other words, the up-regulated expression of circRNAs is a risk factor for NSCLC patients included in these 17 studies (16-18,20-32,34). As shown in Figure 3, the down-regulated expression of circRNAs might be a protected factor for NSCLC patients inversely as the pooled HR of the other 2 studies (19,33) exploring the effect of circRNAs down-regulation to the prognosis of NSCLC patients were 0.45 (95% CI: 0.27–0.73).
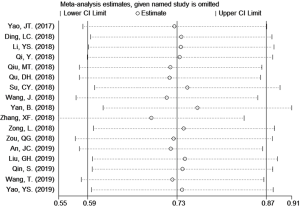
Sensitivity analysis
Sensitivity analysis was applied to test the stability of pooled results. As shown in Figure 4, the pool result of the 17 studies (16-18,20-32,34) in which circRNAs were up-regulated was not significantly affected by removing each eligible study. Regarding to the limited number of the other 2 studies (19,33) with down-regulation of circRNAs, there was no point to perform sensitivity analysis.
Publication bias
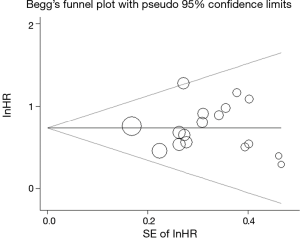
Publication bias was assessed by Begg’s test. As shown in Figure 5, there was no obvious asymmetry in the Begg’s funnel plot; and the P value for Begg’s test was 0.837, which was greater than 0.05. Thus, there was no significant publication bias among the included studies.
Discussion
Research on the functional roles of circRNAs in carcinogenesis is still in its infancy. There is some evidence to indicate that circRNAs could function as valuable biomarkers for cancer diagnosis and prognosis (12). The use of circRNAs as prognostic biomarkers has the following advantages: (I) circRNAs are more stable than linear RNAs; (II) circRNAs are superior to traditional biomarker in terms of organ specificity (35); (III) circRNAs can be easily detected in blood and body fluid (35).
To our knowledge, this meta-analysis is the first time to estimate association between the expression of circRNAs and the prognosis of patients with NSCLC. A total of 16 circRNAs from 19 eligible studies (16-34) comprising 1,650 NSCLC patients were included in this meta-analysis. After a fixed-effects model was performed to estimate pooled HR and the 95% CI (pooled HR =2.08, 95% CI: 1.81–2.40), we became more convinced that up-regulated expression of circRNAs was significantly associated with poor prognosis of patients with NSCLC.
However, there were several limitations in this meta-analysis. Firstly, as HR and 95% CI values from 8 studies (16,17,22,27,31-34) were retrieved from the K-M curves, there could be some marginal error as numerical values were not reported explicitly in the full-text of those studies. Secondly, since we combined multiple types of circRNAs to assess their prognostic role in patients with NSCLC, the heterogeneity might be inevitable. Thirdly, all the included studies of this meta-analysis just tested the expression of circRNAs among Chinese population, thus it is unclear whether our results might be applicable to other populations. Last but not least, we only enrolled 1,650 NSCLC patients from 19 studies (16-34) in this meta-analysis making it hard to widespread the clinical applicability of this meta-analysis because of the small sample size and the small number of studies.
Conclusions
The current meta-analysis might provide evidence that up-regulated expression of circRNAs is significantly associated with poor prognosis of NSCLC patients in Chinese population. Therefore, circRNAs could be novel prognostic biomarkers for NSCLC patients. However, further large-scale prospective studies about the clinical significance of circRNAs are of great need in order to obtain conclusive results.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (NSFC) (No. 81802261).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Reck M, Heigener DF, Mok T, et al. Management of non-small-cell lung cancer: recent developments. Lancet 2013;382:709-19. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. Journal of Hematology & Oncology 2018;11:21. [Crossref] [PubMed]
- Qu S, Yang X, Li X, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015;365:141-8. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384. [Crossref] [PubMed]
- Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res 2015;5:472-80. [PubMed]
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018;37:555. [Crossref] [PubMed]
- Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009;6:e1000097. [Crossref] [PubMed]
- Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [Crossref] [PubMed]
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603-5. [Crossref] [PubMed]
- Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract 2017;213:453-6. [Crossref] [PubMed]
- Ding L, Yao W, Lu J, et al. Upregulation of circ_001569 predicts poor prognosis and promotes cell proliferation in non-small cell lung cancer by regulating the Wnt/beta-catenin pathway. Oncol Lett 2018;16:453-8. [PubMed]
- Li Y, Hu J, Li L, et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun 2018;503:2089-94. [Crossref] [PubMed]
- Liu T, Song Z, Gai Y. Circular RNA circ_0001649 acts as a prognostic biomarker and inhibits NSCLC progression via sponging miR-331-3p and miR-338-5p. Biochem Biophys Res Commun 2018;503:1503-9. [Crossref] [PubMed]
- Qi Y, Zhang B, Wang J, et al. Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene 2018;676:79-85. [Crossref] [PubMed]
- Qiu M, Xia W, Chen R, et al. The Circular RNA circPRKCI Promotes Tumor Growth in Lung Adenocarcinoma. Cancer Res 2018;78:2839-51. [Crossref] [PubMed]
- Qu D, Yan B, Xin R, et al. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res 2018;8:1387-402. [PubMed]
- Su C, Han Y, Zhang H, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-kappa B signalling. J Cell Mol Med 2018;22:3097-107. [Crossref] [PubMed]
- Wang J, Li H. CircRNA circ-0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci 2018;22:3053-60. [PubMed]
- Yan B, Zhang W, Mao XW, et al. Circular RNA ciRS-7 correlates with advance Disease and poor prognosis, and its downregulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2018;22:8712-21. [PubMed]
- Zhang X, Yang D, Wei Y. Overexpressed CDR1 as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther 2018;11:3979-87. [Crossref] [PubMed]
- Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother 2018;102:639-44. [Crossref] [PubMed]
- Zou Q, Wang T, Li B, et al. Overexpression of circ-0067934 is associated with increased cellular proliferation and the prognosis of non-small cell lung cancer. Oncol Lett 2018;16:5551-6. [PubMed]
- An J, Shi H, Zhang N, Song S. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun 2019;511:921-5. [Crossref] [PubMed]
- Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun 2019;513:207-12. [Crossref] [PubMed]
- Qin S, Zhao Y, Lim G, et al. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed Pharmacother 2019;111:244-50. [Crossref] [PubMed]
- Wang T, Du Q, Wu N, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun 2019;513:904-11. [Crossref] [PubMed]
- Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun 2019;508:37-45. [Crossref] [PubMed]
- Yao Y, Hua Q, Zhou Y, et al. CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/beta-catenin signaling pathway. Biomed Pharmacother 2019;111:1367-75. [Crossref] [PubMed]
- Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. Journal of Hematology & Oncology 2018;11:98. [Crossref] [PubMed]