
LncRNA CRNDE modulates cardiac progenitor cells’ proliferation and migration via the miR-181a/LYRM1 axis in hypoxia
Introduction
Myocardial infarction is a severe cardiovascular disease and a significant risk to human life (1). It could cause heart failure and malignant arrhythmia with high morbidities and mortalities (2). Inflammatory responses and cardiomyocyte apoptosis are the most significant outcomes from myocardial infarction (3). Cell apoptosis of cardiomyocytes after myocardial infarction usually causes ventricular remodeling as well as heart failure (4). In this research, we will be focused to explore the mechanisms for cardiac progenitor cell reproduction in myocardial infarction related heart failure.
Long non-coding RNAs (lncRNAs) are a group of RNAs that participate in many kinds of cellular events, such as cell growth, differentiation, and proliferation (5,6). Previous findings suggest that circulating lncRNAs are useful markers for the diagnosis of cardiovascular disease (7,8). For example, several circulating lncRNAs have been discovered as prognostic biomarkers for heart failure, including HOTAIR (9), H19 (10), LIPCR (11), and EGOT (12). The lncRNA CRNDE (colorectal neoplasia differentially expressed gene) is transcribed from chromosome 16 on the strand opposite to the adjacent IRX5 gene (13). LncRNA CRNDE promoted colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling (13). According to Ma et al., CRNDE was involved in the regulation of granulocytic differentiation of acute promyelocytic leukemia (14). But it has not been elucidated whether CRNDE has any effect in the reproduction or migration of cardiac progenitor cells. Inspired by bioinformatic prediction and previous work (13), we are interested in the role of CRNDE in myocardial infarction, and therefore we carried a series of experiments in vitro and in vivo for this investigation.
MicroRNAs (miRNAs, ~24 nucleotides) have been reported to affect the stability and translation of messenger RNAs (15). Many reports have revealed that miRNAs could regulate cell apoptosis, reproduction, development, and differentiation (16). In 2016, Zhu et al. demonstrated the potential of using circulating miR-181a as a novel biomarker for the diagnosis of acute myocardial infarction (17). The expressions of circulating miR-181a in patients with AMI were substantially changed in a time-dependent manner, indicating the value of plasma miR-181a as a novel biomarker for diagnosing MI (17). Herein, we aim to investigate the mysteries of miR-181a and its interactions with CRNDE in myocardial infarction.
According to Qiu, Homo sapiens LYR motif containing 1 (LYRM1) could enhance proliferation and inhibits apoptosis of preadipocytes (18). Zhu et al. reported that LYRM1 increased reproduction and inhibited cell apoptosis during heart development (19). However, its functional mechanism remains to be clarified. In our experiments, we are determined to evaluate the cells’ abilities of proliferation and migration under transfections with LYRM1 over-expression or knockdown. Our experiments, results, and analysis may provide valuable information on its roles in myocardial infarction.
Methods
Cardiac progenitor cell culture
Cardiac progenitor cells were isolated from neonatal adult Sprague-Dawley rats by removing the heart and homogenizing the tissue as described (20). The cardiac progenitor cells were then incubated in DMEM +10% FBS (Gibco, HyClone, USA). After that, cardiac progenitor cells were kept at thirty-seven Celsius and 5% CO2. All procedures on rats are in accordance with the guidelines of the Animal Ethics Committee of The First Affiliated Hospital of Fujian Medical University. All the experiments were conducted according to the principles expressed in the Declaration of Helsinki and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
miRNA and siRNA
si-CRNDE (5'-GTGCTCGAGTGGTTTAAAT-3') and si-LYRM1 (5'-GCAATCATTTCTAGACTAA-3') were made from GenePharma, China. miR-181a-mimic (5'-AACAUUCAACGCUGUCGGUGAGU-3') and miR-181a-inhibitor (5'-ACUCACCGACAGCGUUGAAUGUU-3') were provided by RiboBio, China.
Transfections
The transfections of siRNAs and miRNAs in cardiac progenitor cells were carried out by lipofectamine-2000 (Invitrogen, USA). Prior to transfections, we incubated cardiac progenitor cells in the medium. si-CRNDE or si-LYRM1, and miR-181a-mimic or miR-181a-inhibitor were transfected to the cells with lipofectamine 2000 (Invitrogen, USA).
Quantitative real time-PCR (qRT-PCR)
RNAs were extracted by Trizol (Invitrogen, USA). We made cDNA by EasyScript and SuperMix (Transgen biotech, USA). 10 ng cDNA was prepared for qRT-PCR by SYBR Green in Prism 7500 (Applied Biosystems, Thermo Fisher Scientific, USA). GAPDH and U6 were controls. Table 1 showed the primer sequences.

Full table
Western blotting
The cardiac progenitor cells were lysed, and the proteins were separated through SDS-PAGE and transferred to nitrocellulose membranes (Millipore, USA). The membranes were firstly blocked, treated with anti-LYRM1 (1:500, Abcam), then incubated with horseradish peroxidase linked anti-IgG, and visualized by Chemiluminescence Kit (Thermo Fisher Scientific, USA). Gel-pro was used for statistical analysis.
Luciferase reporter assays
The CRNDE-WT (wild-type), CRNDE-MT (mutant), LYRM1-WT, and LYRM1-MT including the common sequences with miR-181a were sub-cloned to luciferase reporter plasmids. The reporter vectors and miR-181a-mimic or NC-mimic were transfected to cardiac progenitor cells with lipofectamine-2000. After 2 days’ transfections, cardiac progenitor cells were lysed for luciferase activities’ measurement.
CCK-8 assays
A total of 5×104 cells/mL of cardiac progenitor cells after various treatments were grown in 96-well plate for 1 day. Then, cardiac progenitor cells were washed, and the levels of cell growth were evaluated by CCK-8 assays (Transgen, USA). Ten mL CCK-8 solution was placed to every well, and let incubations for four hours at thirty-seven Celsius. A microplate reader (Beckman, USA) was used to measure the absorbance at 450 nm.
Cell migration
Cardiac progenitor cells after various treatments were placed on the top transwell chamber (Corning, USA) with DMEM and 0.5% FBS. The bottom chamber had DMEM and 10% FBS. After 1 day’s incubations, the cardiac progenitor cells on the bottom were cleaned, fixed, and stained by 0.1% crystal violet for fifteen minutes. We counted the migration cells by microscopy (Zeiss, USA).
EdU assays
Cardiac progenitor cells were placed in the 24-well plate (1,500 cells/well). EdU was then pipetted to every well and had incaution for two hours. After fixing the cardiac progenitor cells with 4% paraformaldehyde for half an hour at 25 Celsius, we stained cardiac progenitor cells by Apollo (Ribobio) and detected the signals by fluorescence microscope (Zeiss, USA).
Data analysis
SPSS 17.0 was utilized to analyze the data, shown as mean ± SD. T-test was used for comparisons between two groups. The one-way ANOVA test, followed by Bonferroni’s post-hoc test, was performed to analyze the difference for over two groups. P<0.05 was considered significant.
Results
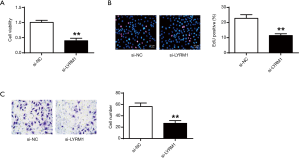
Hypoxia enhanced stem cell proliferation and migration abilities
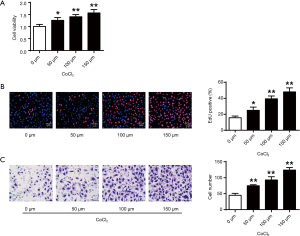
Figure 1A showed that hypoxia could enhance the cell viabilities of cardiac progenitor cells, which was positively correlated with the dose of CoCl2 (P<0.05, P<0.01) (Figure 1A). From Figure 1B,C, the cardiac progenitor cell reproduction and migration potentials were also enhanced in CoCl2-related hypoxia. In consistence with the literature, hypoxia could enhance cell reproduction and migration abilities.
CRNDE affected cardiac progenitor cell proliferation and migration under hypoxic conditions
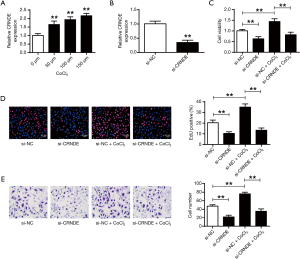
Figure 2A illustrated the qRT-PCR results that CRNDE expressions substantially increased in a CoCl2 dose-dependent manner (P<0.01). Si-CRNDE was made to suppress the expressions of CRNDE in cardiac progenitor cells (P<0.01) (Figure 2B). In Figure 2C, we noticed that the knockdown of CRNDE substantially reduced the cell viabilities of cardiac progenitor cells in hypoxic conditions (P<0.01). Figure 2D,E displayed that the reproduction and migration potentials of cardiac progenitor cells were also suppressed after CRNDE suppression in hypoxia. Obviously, CRNDE could affect cardiac progenitor cell reproduction and migration following hypoxic stimuli.
CRNDE modulated cardiac progenitor cell proliferation and migration via targeting miR-181a in hypoxia
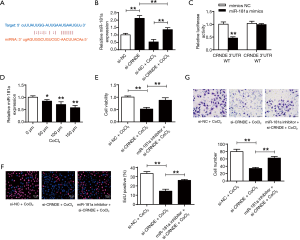
The binding scheme between CRNDE and miR-181a were shown in Figure 3A, with shared binding sequences. According to Figure 3B, CRNDE suppression could greatly elevate miR-181a expressions in both normoxic and hypoxic conditions (P<0.01). Figure 3C showed that miR-181a mimic reduced the luciferase activities of the CRNDE-WT, but didn’t affect the CRNDE-MT (P<0.01). As shown in Figure 3D, the miR-181a expressions were dramatically lowered in cardiac progenitor cells in hypoxia (P<0.05, P<0.01). In addition, the miR-181a-inhibitor substantially restored the cell viabilities (P<0.01) (Figure 3E), and abilities of reproduction (Figure 3F) and migration (Figure 3G) in cardiac progenitor cells. It was illustrated that CRNDE could modulate cardiac progenitor cell proliferation and migration via miR-181a in hypoxic conditions.
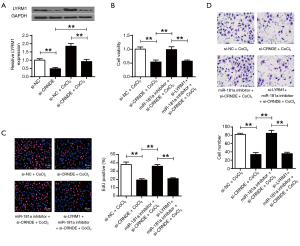
LYRM1 suppressions lowered the protection from miR-181a-inhibitor on cardiac progenitor cells after hypoxia
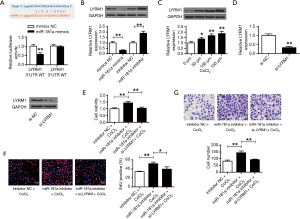
Figure 4A used PicTar and TargetScan to predict that LYRM1 was a target of miR-181a. It shows that miR-181a mimic substantially repressed the luciferase activities of cell transfected with LYRM1-WT (P<0.01). But miR-181a mimic didn’t suppress the luciferase activities of LYRM1-MT. It indicated that LYRM1 was targeted by miR-181a. Figure 4B discovered that LYRM1 expressions were decreased in cardiac progenitor cells after transfecting with miR-181a-mimic but increased in cardiac progenitor cells by miR-181a-inhibitor (P<0.01). Figure 4C showed that LYRM1 expressions were promoted in CoCl2-related hypoxia (P<0.05, P<0.01). Figure 4D illustrated that mRNA and protein levels of LYRM1 were markedly reduced in cardiac progenitor cells after siLYRM1 transfections (P<0.01). However, LYRM1 knockdown in cardiac progenitor cells under hypoxic conditions repressed cell viabilities (Figure 4E), reproduction (Figure 4F) and migration (Figure 4G) abilities that were induced by miR-181a-inhibitor (P<0.01). It indicated that miR-181a modulated cardiac progenitor cell functions though LYRM1.
Si-LYRM1 inhibited CPC viability and migration ability
Then, we assessed the effect of si-LYRM1 on CPCs’ viability and migration ability. According to Figure 5A, si-LYRM1 greatly inhibited the cell viability of CPC, in contrast to the si-NC. From Figure 5B,C, we found that si-LYRM1 also suppressed cell proliferation and migration abilities. It was obvious that the knockdown of LYRM1 inhibited CPC viability and migration ability.
CRNDE modulated CRNDE/miR-181a/LYRM1 in cardiac progenitor cells after hypoxia
According to Figure 6A, the mRNA and the protein levels of LYRM1 were dramatically reduced after CRNDE suppression (P<0.01). It suggested that LYRM1 was positively correlated with CRNDE. In Figure 6B, cell viabilities were suppressed by si-CRNDE but were elevated by miR-181a-inhibitor. But the addition of si-LYRM1 attenuated this effect. Similarly, we found that CRNDE/miR-181a/LYRM1 axis affected cardiac progenitor cell reproduction (Figure 6C) and migration (Figure 6D) abilities under hypoxic conditions (P<0.01). Overall, our results suggested that CRNDE exerted its roles in modulating cardiac progenitor cell reproductions and migration by upregulation of LYRM1 by targeting miR-181a.
Discussion
It was well known that cardiac progenitor cells can quickly lose their reproduction abilities in infarcted myocardium activities (21). CCK-8 assays results found that hypoxia could enhance the cell viabilities of cardiac progenitor cells, which was positively correlated with the dose of CoCl2. The reproduction and migration potentials of cardiac progenitor cells were also enhanced in CoCl2-related hypoxia. Previous reports revealed that the expression levels of lncRNA CRNDE attenuated cardiac fibrosis via Smad3-Crnde negative feedback in diabetic cardiomyopathy (22), and also participated in the regulation of the proliferation and migration of vascular smooth muscle cells (23). The qRT-PCR results that CRNDE expressions substantially increased in a CoCl2 dose-dependent manner. The knockdown of CRNDE considerably reduced the cell viabilities of cardiac progenitor cells in hypoxic conditions. The reproduction and migration potentials of cardiac progenitor cells were also suppressed after CRNDE suppression in hypoxia. In agreements with previous findings, we also found that CRNDE could affect cardiac progenitor cell proliferation and migration by hypoxic stimuli.
MiRNAs were proved to participate in the regulation of a variety of cellular events such as apoptosis, reproduction, development, and differentiation (24,25). For instance, circulating miR-181a as a potential novel biomarker for diagnosis of acute myocardial infarction. Circulating miR-181a levels in patients with MI were substantially changed in a time-dependent manner, indicating the value of plasma miR-181a as a novel biomarker for diagnosing MI (17). Ji et al. demonstrated that CRNDE enhances hepatocellular carcinoma cell proliferation, invasion, and migration via regulating the miR-203/BCAT1 axis (26). It was shown that CRNDE could bind with some specific miRNAs and exert its biological functions in cellular processes (13,26). In our study, CRNDE suppression could much elevate miR-181a expressions, and miR-181a-mimic inhibited the luciferase activities of CRNDE-WT but didn’t affect the CRNDE-MT. Additionally, miR-181a-inhibitor substantially restored the cell viabilities and abilities of reproduction and migration in cardiac progenitor cells following a CRNDE knockdown under hypoxic stimuli. For the first time, we discovered that CRNDE could regulate cardiac progenitor cell proliferation and migration by targeting miR-181a in hypoxic conditions.
According to Chen, knockdown of LYRM1 substantially inhibited reproduction and differentiation and enhanced apoptosis in an embryonic carcinoma cell model of cardiac differentiation (27). In our experiments, LYRM1 expressions were decreased in cardiac progenitor cells by miR-181a mimic but elevated by miR-181a-inhibitor. MiR-181a mimic substantially repressed the luciferase activities of cell transfected with LYRM1-WT. But miR-181a-mimic didn’t suppress the luciferase activities of LYRM1-MT. It is quite possible that miR-181a could bind with LYRM1. What’s more, LYRM1 knockdown in cardiac progenitor cells under hypoxic conditions repressed cell viabilities, proliferation, and migration abilities that were induced by miR-181a-inhibitor. As far as we know, we are the first to report that miR-181a modulated cardiac progenitor cell functions though LYRM1.
Li demonstrated that lncRNA MALAT1 enhanced cardiac progenitor cell reproduction and migration via up-regulating JMJD6 via targeting miR-125 mRNA (28). In a similar pattern, we found that the protein levels of LYRM1 were dramatically reduced after CRNDE suppression, and LYRM1 was positively correlated with CRNDE. In consistence with previous findings, it was confirmed that CRNDE/miR-181a/LYRM1 axis affected cardiac progenitor cell proliferation and migration abilities under hypoxic conditions. CRNDE exerted its roles in modulating cardiac progenitor cell reproduction and migration by the upregulation of LYRM1 via sponging miR-181a.
It is well known that lncRNAs can act as competing endogenous RNA (ceRNA) or molecular sponges to exert “sponge-like” effects on many specific miRNAs, resulting to the downregulation of miRNA levels, upregulation of mRNA levels (target genes), and inhibition of miRNA/mRNA-mediated functions (29,30). In this study, the lncRNA CRNDE functioned as a molecular sponge for miR-181a to modulate the expression of LYRM1 under CoCl2-regulated hypoxia conditions. The upregulated CRNDE induced by hypoxia isolates miR-181a by binding with it, thus reducing the regulation of miR-181a on LYRM1 and promoting the expression of LYRM1, and vice versa.
Conclusions
Our finding demonstrated that CRNDE could modulate cardiac progenitor cell proliferation and migration potentials via the miR-181a/LYRM1 axis in hypoxia. The results may provide a new regulatory mechanism for cardiac progenitor cell reproduction in hypoxia and a new target for MI-related heart failure therapy.
Acknowledgments
Funding: This work was supported by Fujian natural science foundation (2016J01482); Fujian natural science foundation (2018J01369); Fuzhou health and family planning technology innovation platform construction project (2018-S-WP1).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2020.03.22). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures on rats are in accordance with the guidelines of the Animal Ethics Committee of The First Affiliated Hospital of Fujian Medical University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thygesen K, Alpert JS, White HD. Universal definition of myocardial infarction. J Am Coll Cardiol 2007;50:2173-95. [Crossref] [PubMed]
- Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun 2015;6:6779. [Crossref] [PubMed]
- Prabhu SD, Frangogiannis NG. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res 2016;119:91-112. [Crossref] [PubMed]
- Geng HH, Li R, Su YM, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 2016;11:e0151753. [Crossref] [PubMed]
- Liao J, He Q, Li M, et al. LncRNA MIAT: myocardial infarction associated and more. Gene 2016;578:158-61. [Crossref] [PubMed]
- Gast M, Rauch BH, Haghikia A, et al. Long noncoding RNA NEAT1 modulates immune cell functions and is suppressed in early onset myocardial infarction patients. Cardiovasc Res 2019;115:1886-906. [Crossref] [PubMed]
- Li X, Dai Y, Yan S, et al. Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury following acute myocardial infarction. Biochem Biophys Res Commun 2017;491:1026-33. [Crossref] [PubMed]
- Gu M, Zheng A, Tu W, et al. Circulating LncRNAs as novel, non-invasive biomarkers for prenatal detection of fetal congenital heart defects. Cell Physiol Biochem 2016;38:1459-71. [Crossref] [PubMed]
- Gao L, Liu Y, Guo S, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem 2017;44:1497-508. [PubMed]
- Gong LC, Xu HM, Guo GL, et al. Long non-coding RNA H19 protects H9c2 cells against hypoxia-induced injury by targeting microRNA-139. Cell Physiol Biochem 2017;44:857-69. [Crossref] [PubMed]
- Li M, Wang LF, Yang XC, et al. Circulating long noncoding RNA LIPCAR acts as a novel biomarker in patients with ST-segment elevation myocardial infarction. Med Sci Monit 2018;24:5064. [Crossref] [PubMed]
- Zhang C, Pan S, Aisha A, et al. Recombinant human brain natriuretic peptide regulates PI3K/AKT/mTOR pathway through lncRNA EGOT to attenuate hypoxia-induced injury in H9c2 cardiomyocytes. Biochem Biophys Res Commun 2018;503:1186-93. [Crossref] [PubMed]
- Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017;16:9. [Crossref] [PubMed]
- Ma X, Zhang W, Zhang R, et al. Overexpressed long noncoding RNA CRNDE with distinct alternatively spliced isoforms in multiple cancers. Front Med 2019;13:330-43. [Crossref] [PubMed]
- Yang Y, Cheng HW, Qiu Y, et al. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res 2015;117:450-9. [Crossref] [PubMed]
- Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol 2015;12:135. [Crossref] [PubMed]
- Zhu J, Yao K, Wang Q, et al. Circulating miR-181a as a potential novel biomarker for diagnosis of acute myocardial infarction. Cell Physiol Biochem 2016;40:1591-602. [Crossref] [PubMed]
- Qiu J, Gao CL, Zhang M, et al. LYRM1, a novel gene promotes proliferation and inhibits apoptosis of preadipocytes. Eur J Endocrinol 2009;160:177-84. [Crossref] [PubMed]
- Zhu C, Liu YQ, Chen FK, et al. LYRM1, a gene that promotes proliferation and inhibits apoptosis during heart development. Molecules 2010;15:6974-82. [Crossref] [PubMed]
- Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003;114:763-76. [Crossref] [PubMed]
- Khan M, Nickoloff E, Abramova T, et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res 2015;117:52-64. [Crossref] [PubMed]
- Zheng D, Zhang Y, Hu Y, et al. Long noncoding RNA Crnde attenuates cardiac fibrosis via Smad3‐Crnde negative feedback in diabetic cardiomyopathy. FEBS J 2019;286:1645-55. [Crossref] [PubMed]
- Zhou Y, He X, Liu R, et al. LncRNA CRNDE regulates the proliferation and migration of vascular smooth muscle cells. J Cell Physiol 2019. [Epub ahead of print]. [PubMed]
- Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun 2018;495:2369-75. [Crossref] [PubMed]
- Cheng Y, Xiang G, Meng Y, et al. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod Biol 2016;16:225-33. [Crossref] [PubMed]
- Ji D, Jiang C, Zhang L, et al. LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation, invasion, and migration through regulating miR‐203/BCAT1 axis. J Cell Physiol 2019;234:6548-60. [Crossref] [PubMed]
- Chen YM, Li X, Song GX, et al. Effect of LYRM1 knockdown on proliferation, apoptosis, differentiation and mitochondrial function in the P19 cell model of cardiac differentiation in vitro. J Bioenerg Biomembr 2016;48:33-41. [Crossref] [PubMed]
- Li L, Wang Q, Yuan Z, et al. LncRNA-MALAT1 promotes CPC proliferation and migration in hypoxia by up-regulation of JMJD6 via sponging miR-125. iochem Biophys Res Commun 2018;499:711-8.
- Zhou M, Diao Z, Yue X, et al. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget 2016;7:56383. [Crossref] [PubMed]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Long Non-Coding RNAs. Springer, 2016:271-86.


