
Comprehensive analysis of lncRNA-associated competing endogenous RNA network and immune infiltration in idiopathic pulmonary fibrosis
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic progressive pulmonary interstitial disease with unknown causes, which is the most common pulmonary pathological feature of idiopathic interstitial pneumonia. Although the overall incidence of the disease is low, it tends to increase over time (1). In Europe and North America, there are an estimated eight to 18 cases per 100,000 people per year. Rates are lower in Asia and South America, where they range from 0.5 to 4.2 per 100,000 people per year (2). The median age of IPF diagnosis was 65 years old, and patients younger than 50 years old were rare, with more male patients than female patients (3). The disease progresses insidiously and unpredictably, with a median survival time of 2–4 years after diagnosis (4).
IPF was once thought to be a disease caused by immune disorders, but the clinical application of anti-inflammatory drugs has not improved the prognosis of patients and may even increase the mortality (5,6). A large number of recent studies have shown that the disease is caused by a combination of factors such as genetic factors, environmental risk factors and exposure (7). The basic pathological changes are as follows: the slight injury of lung tissue causes the interaction between epithelial fibroblasts, and induces the activation of storm-producing myofibroblasts, resulting in a large amount of extracellular matrix and the remodeling of lung interstitium (8,9). The clinical manifestations of IPF are primarily exertional dyspnea with or without a dry cough, which is usually not specific and is not easily distinguished from interstitial lung disease in general. So, the main diagnostic challenge is to exclude other types of interstitial lung disease in the cases considered IPF. In terms of disease treatment, numerous multi-center studies have shown the ineffectiveness and toxicity of some conventional drugs, such as prednisolone and azathioprine. At present, nintedanib and pirfenidone have been widely used (10,11), both of which have shown definite effects on extending the survival period, but still have relatively high probability of side effects (12).
Long non-coding RNA (lncRNA) is a type of RNA transcripts, which has been one of the hottest research fields in the life science field for nearly 10 years. It turns out that the human body produces far more non-coding RNA than coding RNA (13). LncRNA is more than 200 nucleotide sequences in length. Although it does not have the function of coding protein, more and more studies have found in recent years that lncRNA plays an important role in tumor, infection, cardiovascular disease, diabetes, lung disease and other extensive diseases (14,15). It is generally believed that lncRNA functions by regulating the binding of microRNA to its target mRNA. However, the characteristics and functions of most lncRNAs have not been elucidated up to now. At present, although some studies have found that some microRNAs are involved in the pathogenesis of IPF (16), there are few studies on the role of lncRNAs in this regard. Limited studies have reported that two lncRNAs, CD99P1 and n341773 may be involved in the regulation of the proliferation and mutation process of pulmonary fibrosis in IPF (17). We expect that the study of lncRNAs involving IPF will provide a new direction for the treatment of IPF.
The immune system plays an intricate role in the pathogenesis of IPF. Previous studies showed that T cells play a beneficial role in pulmonary fibrosis. T cells could induce apoptosis in myofibroblasts from lungs resolving fibrosis (18). Another study assessed the interaction between immune T cells and lung myofibroblasts. The findings revealed that myofibroblasts possess Fas/FasL-pathway-dependent characteristics that enable them to avoid immune surveillance and resulting lung fibrosis (19). Due to technical constraints, conventional immunological infiltration measurement methods cannot comprehensively evaluate the immune effects of different cell types, usually including only one or two cell types. CIBERSORT is a newly developed bioinformatics method for enumeration of immune cells from gene expression profiles (20). This algorithm was successfully validated by flow cytometry and conducted in several types of human cancers such as breast (21), colon (22) and osteosarcoma (23). By using CIBERSORT method, previous research has studied the relationship between immune cells and IPF outcome based on whole blood datasets (24). The study suggested resting memory T cells and monocytes proportions were correlated with IPF prognosis. However, to date, there are no studies investigating the fractions of 22 immune cell types in lung tissues of IPF.
The aims of this study were two-fold: (I) to establish a lncRNA-miRNA-mRNA ceRNA network of IPF and improve our understanding of the mechanism of lncRNAs in IPF; (II) to estimate the proportion of immune cells in IPF lung tissues and assess their relationship with disease severity. Through our research, we hope to further understand the immune mechanism of the disease process and discover possible key targets, so as to make contributions to the improvement of the accuracy of IPF diagnosis and the development of new drugs.
Methods
Data processing
The raw microarray data were retrieved from the Genome Expression Omnibus (GEO) datasets under accession numbers GSE32537 (25) and GSE110147 (26). Both datasets came from the same platform (GPL6244). Lung tissue samples from 141 IPF patients and 61 non-diseased controls were included in the present study. Background correction, normalization and summarization of expression levels were applied using the robust multi-array average method (27). We utilized surrogate variable analysis package to remove batch effects (28). If multiple probes corresponded to a gene symbol, we calculated the average value of the probes and converted the probe names into gene names. In this study, data processing and analysis were carried out using R packages and Perl script. Ethics approval was not required as our data were downloaded directly from public datasets.
Differential expression analysis
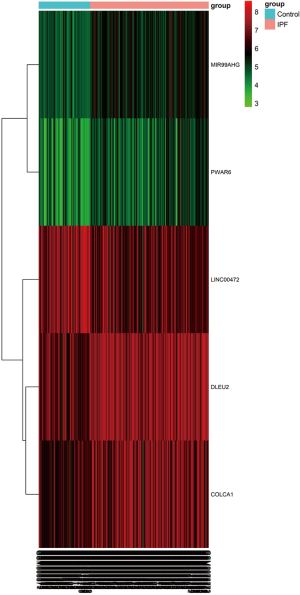
The differentially expressed lncRNAs (DElncRNAs) and mRNAs (DEmRNAs) between IPF and control lung tissue samples were screened by limma package (29). |logFC(fold change)| >0.5 and P value <0.05 were considered significant. The heatmap of DElncRNAs expression was generated using pheatmap packages.
Establishment of the lncRNA–miRNA–mRNA ceRNA network
The competitive endogenous RNA (ceRNA) network was established according to the interactions between DElncRNA, miRNA, and DEmRNA. The DElncRNA–miRNA pairs were predicted using the miRcode database (30), miRDB (31), miRTarBase (32) and TargetScan (33) databases were applied to predict the miRNA targeted mRNAs. MRNAs in all three databases were selected, and those that also belong to DEmRNAs were selected again as the targeted DEmRNAs of miRNA in ceRNA network. The ceRNA network was built and visualized by Cytoscape software (34).
Functional enrichment analysis
To explore the biological processes of DEmRNAs in the ceRNA network, functional enrichment analysis was carried out using Metascape (35). The top 20 most significantly enriched pathways were selected.
Immunity cells analysis
CIBERSORT was applied to calculate immune cell composition from gene expression profiles. Leukocyte signature matrix is the signature genes file which contains 547 genes that distinguish 22 immune cell types. This file together with normalized gene expression data served as input files for CIBERSORT analysis. The number of permutations was set to 1,000 and samples with a CIBERSORT P<0.05 were chosen for further analysis. The heat map of 22 immune cells was generated via the pheatmap package in R. The violin plot showed the ratio of different immune cell types between IPF group and control group, which was performed using the R vioplot package. The correlation between immune cells proportion and clinical factors was analyzed with Wilcoxon test.
Results
Identification of differentially expressed lncRNAs and mRNAs
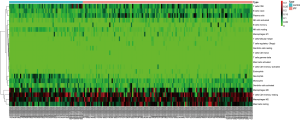
Based on the screening criteria, we obtained the DElncRNAs and DEmRNAs in IPF compared with normal lung tissue samples. A total of five DElncRNAs (four up-, and one down-regulated) and 1,688 DEmRNAs (1,010 up- and 678 down-regulated) were identified. The heatmap of DElncRNAs was shown in Figure 1.
Construction of ceRNA networks and functional analysis
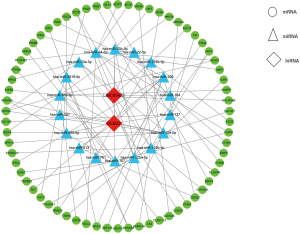
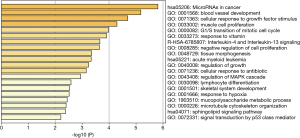
The two DElncRNAs were acquired from the miRcode database, and 145 lncRNA-miRNA interactions were obtained. We observed these 145 miRNAs targeted 749 mRNAsin miRDB, miRTarBase and TargetScan databases. Among the 749 targeted mRNAs, only 66 mRNAs were found in the 1,688 DEmRNAs. The lncRNA-miRNA-mRNA ceRNA network was built according to the above data. As shown in Figure 2, the network was composed of two DElncRNAs, 18 miRNAs, 66 DEmRNAs and 100 edges. The key DElncRNAs in the entire network were DLEU2 and LINC00472, which may play a significant role in the pathogenesis of IPF. Moreover, the DEmRNAs in the ceRNA network were mainly involved in MicroRNAs in cancer, blood vessel development and cellular response to growth factor stimulus (Figure 3).
Immunity cells analysis
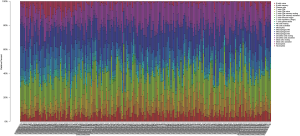
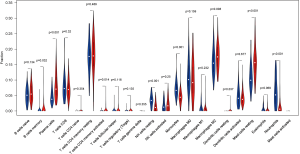
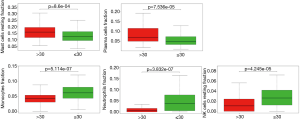
We applied the CIBERSORT algorithm to perform 1,000 permutations and obtained all samples with a P value less than 0.05. The proportion of 22 immune cells in all samples was shown in Figure 4. Resting memory T cells CD4, M2 Macrophages, resting Mast cells and M0 Macrophages comprised the majority of immune cells. As shown in Figure 5, important differences were found between IPF lung samples and control lung samples. The violin plot (Figure 6) showed that IPF samples had a higher proportion of plasma cells, M2 macrophages, resting mast cells and a lower proportion of resting NK cells, monocytes, neutrophils compared with control samples. This study investigated the correlation between 22 distinct immune cell types and clinical traits. There was no correlation between gender and the proportion of immune cells. The proportion of plasma cells and resting mast cells were found to be positively correlated with the St. George’s Respiratory Questionnaire (SGRQ) total score while neutrophils, monocytes, resting NK cells appeared to be negatively correlated with the SGRQ total score (Figure 7). These results indicated that immune cells were associated with the severity of IPF.
Discussion
IPF is a severe life-threatening disease in global population that generally leads to death within a few years after diagnosis. The molecular pathogenesis of IPF remains not fully understood. While a few studies have indicated that lncRNAs play key roles in the progression of IPF (17), no study has constructed lncRNA-associated ceRNA network in IPF.
In the present study, we used gene expression data from the GEO database to screen significantly expressed lncRNAs and mRNAs in IPF. A total of five lncRNAs and 1,688 mRNAs were significantly altered in IPF compared with control groups. Then, we established the lncRNA-miRNA-mRNA ceRNA network, which included 2 lncRNAs, 66 mRNA and 18 miRNAs. Two novel candidate lncRNAs were detected, of which DLEU2 was upregulated and LINC00472 was downregulated in IPF. DLEU2 is a cancer-related lncRNA with 15 exons and several transcripts (36). Some studies reported that DLEU2 was significantly upregulated in various cancers, including gastric cancer and pancreatic ductal adenocarcinoma (37,38). Besides, DLEU2 may serve as a potential prognostic biomarker for esophageal adenocarcinoma (39). DLEU2 may have oncogenic functions and modulate the proliferation, migration and invasion of cancer. Our study suggested that overexpression of DLEU2 might promote the progression of IPF. Studies on DLEU2 were very limited and levels of DLEU2 in IPF have not been previously discussed. Consequently, future studies are warranted to confirm its function in IPF. Previous studies have reported that LINC00472 was found to be downregulated in various human cancers and lower expression of LINC00472 was associated with worse prognosis (40). LINC00472 inhibited cell proliferation and migration in many kinds of human cancers, suggesting that LINC00472 may be an important tumor suppressor. Moreover, LINC00472 was significantly downregulated in diabetic kidney disease patients compared to the control samples (41). These studies suggested that LINC00472 may play a crucial role in human diseases. However, very little is known about the role of LINC00472 in IPF. Further studies are required to elucidate the function of LINC00472 in IPF.
To better understand the function of ceRNA networks in IPF, functional enrichment analysis of differentially expressed mRNAs was performed using Metascape. The results indicated that the DEmRNAs mainly involved in MicroRNAs in cancer, blood vessel development and cellular response to growth factor stimulus. Many dysregulated microRNAs were commonly involved in the molecular pathogenesis of IPF and cancers, especially lung cancer (42). IPF and lung cancer showed similar risk factors, pathogenic pathways, genetic changes and worse prognosis (43). our results indicated that not only lncRNAs in IPF ceRNA network were cancer-related but also mRNAs were involved in pathways crucial to cancers. All the evidence above showed that IPF can be considered a cancer-like disease, which can help in better understanding the pathogenesis of IPF. Importantly, viewing IPF as a cancer-like disorder might attract more public attention and draw lessons from the experience of cancer treatment and encourage the use of cancer drugs for IPF treatment (44).
In this paper, CIBERSORT was applied to estimate the fraction of 22 immune cells in IPF and controls lung tissue samples. We found that the proportions of plasma cells, M2 Macrophages and resting mast cells in IPF lung tissues were higher than control samples; on the contrary, the proportions of resting NK cells, monocytes and neutrophils in IPF lung tissue samples were lower than control samples. In addition, we investigated the relationship between immune cells and clinical traits. The proportions of plasma cells and resting mast cells were positively correlated with the SGRQ total score while neutrophils, monocytes, resting NK cells were negatively correlated with the SGRQ total score. This suggested that immune cells in lung tissue samples might predict disease severity and mortality in IPF. Plasma cells, also known as effector B cells, are derived from B cells and participate in humoral immune response. Abnormalities of B cell activating factor and CD20+ B cells had been reported for IPF lung tissues (45,46). Moreover, B cell targeted therapies showed improvement of pulmonary function and alleviation of pulmonary fibrosis for IPF patients (47). These data suggested plasma cells may play a key role in the pathogenesis of IPF. Mast cells play essential roles in both innate and adaptive immunity. Increased numbers of mast cells were reported in the alveolar parenchyma in patients with IPF (48). Our results showed that resting mast cells, not activated mast cells were significantly increased in IPF lung tissues and positively correlated with the SGRQ total score. The role of resting mast cells in IPF lung tissues may be a significant research area in identifying new anti-IPF therapeutic strategies. Neutrophils can regulate the production of cytokines and turnover of extracellular matrix, which may contribute to pulmonary fibrosis (49). Moreover, neutrophile lastase levels was higher in bronchoalveolar lavage fluid of IPF patients (50) and the expression of neutrophil chemoattractant, IL-8, was increased in IPF (51). One previous study reported that neutrophil counts in bronchoalveolar lavage fluid were not higher than normal controls in IPF patients (52). Also, our study showed that neutrophils were significantly decreased in IPF lung tissues compared with control lung tissues. The role of neutrophils in IPF has remained controversial and further studies will be required to understand its role in IPF pathogenesis. To date, few literatures have studied the proportions of monocytes and resting NK cells in IPF lung tissues. Future efforts should be made to understand their relevance to IPF.
Conclusions
Taken together, this is the first study to construct lncRNA-miRNA-mRNA ceRNA network of IPF. Two lncRNA identified in the ceRNA network may serve as novel biomarkers and therapeutic targets. The ceRNA network will help us to better understand the pathogenesis of IPF. In addition, this is the first study to estimate the proportions of immune cells in lung tissues of IPF. We found that immune cells in lung tissues may predict disease severity and participate in the development of IPF. Future prospective researches are required to confirm the findings of the current study.
Acknowledgments
We are grateful to GSE32537 and GSE110147 from Genome Expression Omnibus datasets.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-19-2842). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Xaubet A, Ancochea J, Molina-Molina M. Idiopathic pulmonary fibrosis. Med Clin (Barc) 2017;148:170-5. [Crossref] [PubMed]
- Hutchinson J, Fogarty A, Hubbard R, et al. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. Eur Respir J 2015;46:795-806. [Crossref] [PubMed]
- Raghu G, Weycker D, Edelsberg J, et al. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2006;174:810-6. [Crossref] [PubMed]
- Ley B, Collard HR, King TE Jr. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2011;183:431-40. [Crossref] [PubMed]
- Selman M, King TE, Pardo A. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 2001;134:136-51. [Crossref] [PubMed]
- Raghu G, Anstrom KJ, King TE Jr, et al. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med 2012;366:1968-77. [Crossref] [PubMed]
- Martinez FJ, Collard HR, Pardo A, et al. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers 2017;3:17074. [Crossref] [PubMed]
- Rock JR, Barkauskas CE, Cronce MJ, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A 2011;108:E1475-83. [Crossref] [PubMed]
- Hung C, Linn G, Chow YH, et al. Role of lung pericytes and resident fibroblasts in the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med 2013;188:820-30. [Crossref] [PubMed]
- Richeldi L, du Bois RM, Raghu G, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med 2014;370:2071-82. [Crossref] [PubMed]
- King TE Jr, Bradford WZ, Castro-Bernardini S, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med 2014;370:2083-92. [Crossref] [PubMed]
- Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet 2017;389:1941-52. [Crossref] [PubMed]
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484-8. [Crossref] [PubMed]
- Henderson J, Distler J, O'Reilly S. The Role of Epigenetic Modifications in Systemic Sclerosis: A Druggable Target. Trends Mol Med 2019;25:395-411. [Crossref] [PubMed]
- O'Reilly S. Epigenetics in fibrosis. Mol Aspects Med 2017;54:89-102. [Crossref] [PubMed]
- Liu G, Friggeri A, Yang Y, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2010;207:1589-97. [Crossref] [PubMed]
- Huang C, Yang Y, Liu L. Interaction of long noncoding RNAs and microRNAs in the pathogenesis of idiopathic pulmonary fibrosis. Physiol Genomics 2015;47:463-9. [Crossref] [PubMed]
- Wallach-Dayan SB, Golan-Gerstl R, Breuer R. Evasion of myofibroblasts from immune surveillance: a mechanism for tissue fibrosis. Proc Natl Acad Sci U S A 2007;104:20460-5. [Crossref] [PubMed]
- Wallach-Dayan SB, Elkayam L, Golan-Gerstl R, et al. Cutting edge: FasL(+) immune cells promote resolution of fibrosis. J Autoimmun 2015;59:67-76. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Ali HR, Chlon L, Pharoah PD, et al. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med 2016;13:e1002194. [Crossref] [PubMed]
- Xiong Y, Wang K, Zhou H, et al. Profiles of immune infiltration in colorectal cancer and their clinical significant: A gene expression-based study. Cancer Med 2018;7:4496-508. [Crossref] [PubMed]
- Yang X, Zhang W, Xu P. NK cell and macrophages confer prognosis and reflect immune status in osteosarcoma. J Cell Biochem 2018. [Epub ahead of print]. [PubMed]
- Liu YZ, Saito S, Morris GF, et al. Proportions of resting memory T cells and monocytes in blood have prognostic significance in idiopathic pulmonary fibrosis. Genomics 2019;111:1343-50. [Crossref] [PubMed]
- Yang IV, Coldren CD, Leach SM, et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax 2013;68:1114-21. [Crossref] [PubMed]
- Cecchini MJ, Hosein K, Howlett CJ, et al. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir Res 2018;19:153. [Crossref] [PubMed]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249-64. [Crossref] [PubMed]
- Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882-3. [Crossref] [PubMed]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3.
- Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012;28:2062-3. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239-47. [Crossref] [PubMed]
- Fromm B, Billipp T, Peck LE, et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu Rev Genet 2015;49:213-42. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Tripathi S, Pohl MO, Zhou Y, et al. Meta- and Orthogonal Integration of Influenza "OMICs" Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015;18:723-35. [Crossref] [PubMed]
- Corcoran MM, Hammarsund M, Zhu C, et al. DLEU2 encodes an antisense RNA for the putative bicistronic RFP2/LEU5 gene in humans and mouse. Genes Chromosomes Cancer 2004;40:285-97. [Crossref] [PubMed]
- Liu H, Zhang Z, Wu N, et al. Integrative Analysis of Dysregulated lncRNA-Associated ceRNA Network Reveals Functional lncRNAs in Gastric Cancer. Genes (Basel) 2018. [Crossref] [PubMed]
- Giulietti M, Righetti A, Principato G, et al. LncRNA co-expression Network Analysis Reveals Novel Biomarkers for Pancreatic Cancer. Carcinogenesis 2018;39:1016-25. [Crossref] [PubMed]
- Ma W, Zhang CQ, Dang CX, et al. Upregulated long-non-coding RNA DLEU2 exon 9 expression was an independent indicator of unfavorable overall survival in patients with esophageal adenocarcinoma. Biomed Pharmacother 2019;113:108655. [Crossref] [PubMed]
- Wang J, Zhang C, He W, et al. Construction and comprehensive analysis of dysregulated long non-coding RNA-associated competing endogenous RNA network in clear cell renal cell carcinoma. J Cell Biochem 2018. [Epub ahead of print]. [PubMed]
- Wang YZ, Zhu DY, Xie XM, et al. EA15, MIR22, LINC00472 as diagnostic markers for diabetic kidney disease. J Cell Physiol 2019;234:8797-803. [Crossref] [PubMed]
- Mizuno K, Mataki H, Seki N, et al. MicroRNAs in non-small cell lung cancer and idiopathic pulmonary fibrosis. J Hum Genet 2017;62:57-65. [Crossref] [PubMed]
- Vancheri C. Idiopathic pulmonary fibrosis and cancer: do they really look similar? BMC Med 2015;13:220. [Crossref] [PubMed]
- Vancheri C. Common pathways in idiopathic pulmonary fibrosis and cancer. Eur Respir Rev 2013;22:265-72. [Crossref] [PubMed]
- François A, Gombault A, Villeret B, et al. B cell activating factor is central to bleomycin- and IL-17-mediated experimental pulmonary fibrosis. J Autoimmun 2015;56:1-11. [Crossref] [PubMed]
- Todd NW, Scheraga RG, Galvin JR, et al. Lymphocyte aggregates persist and accumulate in the lungs of patients with idiopathic pulmonary fibrosis. J Inflamm Res 2013;6:63-70. [Crossref] [PubMed]
- Keir GJ, Maher TM, Ming D, et al. Rituximab in severe, treatment-refractory interstitial lung disease. Respirology 2014;19:353-9. [Crossref] [PubMed]
- Andersson CK, Andersson-Sjoland A, Mori M, et al. Activated MCTC mast cells infiltrate diseased lung areas in cystic fibrosis and idiopathic pulmonary fibrosis. Respir Res 2011;12:139. [Crossref] [PubMed]
- Desai O, Winkler J, Minasyan M, et al. The Role of Immune and Inflammatory Cells in Idiopathic Pulmonary Fibrosis. Front Med (Lausanne) 2018;5:43. [Crossref] [PubMed]
- Yamanouchi H, Fujita J, Hojo S, et al. Neutrophil elastase: alpha-1-proteinase inhibitor complex in serum and bronchoalveolar lavage fluid in patients with pulmonary fibrosis. Eur Respir J 1998;11:120-5. [Crossref] [PubMed]
- Xaubet A, Agusti C, Luburich P, et al. Interleukin-8 expression in bronchoalveolar lavage cells in the evaluation of alveolitis in idiopathic pulmonary fibrosis. Respir Med 1998;92:338-44. [Crossref] [PubMed]
- Obayashi Y, Yamadori I, Fujita J, et al. The role of neutrophils in the pathogenesis of idiopathic pulmonary fibrosis. Chest 1997;112:1338-43. [Crossref] [PubMed]


