
Applications of cell-free circulating tumor DNA detection in EGFR mutant lung cancer
EGFR as a model of non-invasive detection monitoring
The assessment of molecular alterations for patients with non-squamous non-small cell lung cancer (NSCLC) has been adopted into guidelines to determine suitability for targeted treatments (1,2). Tissue adequacy in detecting resistance mutations is an important reason to consider plasma testing. Up to 30% of NSCLC patients cannot provide sufficient tumor samples for molecular testing at diagnosis or progression (3). Circulating tumor DNA serves as an alternative method to non-invasively detect molecular aberrations in NSCLC when tissue is not available (4) (Figure 1).
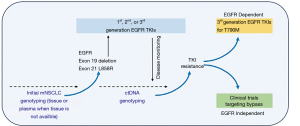
Plasma detection of EGFR mutations serves as a model to demonstrate the clinical utility of a non-invasive testing study for EGFR-TKI selection in the era of precision medicine (5). The therascreen (Qiagen) EGFR RGQ PCR Kit and cobas® EGFR Mutation Test v2 are qPCR assays approved by the EMA and FDA, respectively, for the analysis of plasma ctDNA (6). The cobas® EGFR Mutation Test (v1) is a qualitative real-time polymerase chain reaction (RT-PCR) test approved in 2013 for exon 19 del and L858R substitution mutations in FFPE NSCLC tissue specimens (7). The FDA approved the cobas® EGFR Mutation Test v2 in 2015 for both FFPE and plasma specimens and includes the assessment of T790M mutations, exon 18 substitutions, exon 19 deletions, and exon 20 and 21 substitutions when tissue is not available (7,8). There have been several plasma based laboratory developed tests that utilize next generation sequencing (NGS) strategies, and these are currently pending regulatory approvals.
Initial genotyping with PCR technologies
The need to initially genotype patients with prompt turn-around times is fundamentally important in therapeutic decision-making. Basing front-line approaches on the results of a patient’s molecular profile has maximized efficacy and mitigated potential toxicity with sequential treatments. The overall survival of patients on targeted therapies has superseded historical controls without targeted approaches.
Among Japanese NSCLC patients on first-line gefitinib, 48% patients had EGFR exon 19 deletions and L858R mutations, and the identification of these mutations predicted response to gefitinib (9). In another study of forty-two patients receiving gefitinib, EGFR mutation detected by ctDNA was predictive of improved objective response rate (P=0.001) and median progression free survival (P=0.044) to gefitinib (10).
In a recent prospective trial called the Non-invasive vs. Invasive Lung Evaluation study (NILE), a hybridization captures targeted sequencing NGS assay detected FDA-approved mutations (EGFR, ALK, ROS1, BRAF) in previously untreated patients with high concordance of driver alleles compared to tissue genotyping tests (>98.2%). The utilization of cfDNA combined with tissue increased detection by 32% in patients with negative, not accessible, or insufficient tissue and median turn around time of 9 days (11). Another PCR based analysis showed a 91% overall concordance of EGFR mutations detected in plasma and tissue in 196 NSCLC patients studied (12). A high correlation was seen in a separate study with the EGFR gene as detected in plasma versus those detected in the matched tumor sample (87.7% concordance) (13). There have been earlier studies that have lacked this high level of concordance with potential underlying variables including differences in assay platforms, spatial and temporal factors in tumor burden, tumor heterogeneity, interval treatment, potential germline DNA contamination, and variants which may derive from clonal hematopoiesis (14).
The FASTACT-2 trial demonstrated the predictive value of serial ctDNA EGFR testing by exploring prospectively whether baseline tissue biopsy correlates with Cobas® EGFR ctDNA testing. Analysis of circulating tumor DNA was performed in patients with stage IIIB/IV NSCLC randomized to receive chemotherapy followed by maintenance with either erlotinib or placebo (15). A planned retrospective analysis of advanced NSCLC treated first-line with chemotherapy and erlotinib showed an 88% concordance between blood and tissue testing with a sensitivity and specificity of 75% and 96%, respectively. Median PFS for ctDNA was 13.1 months for EGFR mutant ctDNA-positive patients treated with erlotinib, and 6 months for patients treated with placebo. For EGFR wild-type ctDNA patients, erlotinib or placebo yielded similar survival (~6 months). Patients with EGFR mutant ctDNA detected at baseline who experienced a complete elimination at the end of an induction period of treatment had improved progression-free survival (12 vs. 7.2 months) and overall survival (31.9 vs. 18.2 months) (15,16).
Some additional studies have evaluated whether the upfront ctDNA load may correlate with clinical outcome endpoints. The EURTAC176 clinical trial tested ctDNA as a proxy for tissue EGFR testing with an RT-PCR (TaqMan) assay to assess for EGFR mutations. Patients with EGFR mutations detected in pre-treatment ctDNA predicted shorter OS in univariate analysis. Tissue confirmed L858R mutation when detected in plasma correlated with a median OS of 13.7 months compared to 27.7 months for those with the mutation undetectable in cfDNA (HR 2.22) (17).
Tracking tumor burden with NGS based approaches
A number of studies have demonstrated that ctDNA load can correlate with changes in tumor burden as detected with ultrasensitive NGS approaches. Newman et al. evaluated the effectiveness of CAPP-Seq for minimal residual disease (MRD) detection and monitoring using plasma samples from healthy controls and a NSCLC cohort (18). In patients with Stage I-IV NSCLC, ctDNA was detected in all patients, in half of the patients with stage I and with a 96% mutant allele fraction specificity and a detection threshold of ~0.02%. Chabon et al. serially tracked EGFR tumor load and resistance mutations on therapy with rises that were concordant with CT scan tumor increases (19). Chaudhuri et al. evaluated CAPP-Seq in over 250 samples from forty patients being treated with curative intent diagnosed with stage I-III NSCLC and in fifty-four healthy controls. Plasma circulating DNA was detected post-treatment in the vast majority (94%) of patients having recurrence. ctDNA detection after treatment preceded progression seen on imaging modalities in about a third of patients by approximately 5 months. About half of the patients had actionable mutations in plasma, providing an opportunity for personalized adjuvant treatment (20). A novel technology called TEC-SEQ (Targeted Error Correction Sequencing) has identified genomic aberrations in lung cancer in stage I and II patients at a rate of approximately 45–50% (21). This strategy facilitates early MRD monitoring and has been utilized for tracking response to therapy (22). Ongoing studies are using fragment length to characterize tumor specific DNA from wild type DNA (23).
Resistance monitoring
Circulating tumor DNA has been used as a companion diagnostic to monitor for the EGFR gatekeeper T790M mutation after treatment with first- and second-generation inhibitors (24,25). Oxnard et al. showed similar efficacy outcomes with osimertinib with plasma digital PCR by BEAMing or tissue T790M testing (26). The positive predictive value (PPV) of ctDNA was 100% for L858R and EGFR 19 deletions, and 79% for T790M mutations. The study had a 30% false negative rate for plasma genotyping. Patients with a negative plasma T790M result have necessitated a primary tumor biopsy to confirm the absence of T790M.
In a separate retrospective study of EGFR mutant patients, BEAMing digital PCR was able to detect 70% T790M mutants. About a third (31%) of the patients with T790M-negative tumors had T790M positive ctDNA. In this study, response rates and PFS correlated with plasma positive T790M or tissue positive T790M (27). In another trial, an association between ctDNA load and OS was seen, whereas no correlation was noted with serial PET/CT tumor volume or avidity. Increased ctDNA independently associated with a shorter overall survival (28).
Tumor heterogeneity and acquired resistance
Co-occurring alterations or tumoral heterogeneity may explain drug resistance to EGFR inhibitors. Plasma ctDNA sampling can provide a comprehensive analysis across metastatic sites when detectable (6,29). Furthermore, a tumoral molecular profile may evolve dynamically over time as a result of selection pressures on therapy (30). Understanding the evolution of selective pressures contributing to resistance mutations may help guide sequential targeted therapy (31-34). Third generation EGFR tyrosine kinase inhibitors irreversibly inhibit EGFR T790M resistance and have received front-line approval in the metastatic setting for patients with EGFR mutation (35). Patients treated with osimertinib whom relapse may acquire new genetic alterations including the EGFR C797S mutation (36). A variety of additional alterations have been observed and may include KRAS mutations, BRAF V600 mutations, HER2 amplification, and MET amplification among others (35-37).
In patients with NSCLC treated with third generation EGFR TKIs, acquired EGFR C797S mutation has been identified on serial cfDNA specimens (38). Analysis of plasma collected from 15 patients treated with osimertinib detected three molecular subtypes emerging at resistance defined by EGFR C797S or a bypass mechanism (39). In a subset of patients who had progressed on rociletinib, different EGFR activating mutations (i.e., L798I) and bypass pathways with MET amplification have been detected in ctDNA and track with tumor resistance (19).
Early markers of response
A number of studies have shown that the dynamic monitoring of ctDNA load can provide insights into the detection of treatment response (19,40). Shorter PFS and OS can be associated with early ctDNA detection independent of confounding factors including age, stage, and histological subtype (41,42). We have employed an ultrasensitive liquid biopsy approach (TEC-SEQ) to serially evaluate patients with advanced NSCLC who have received tyrosine kinase inhibitors including erlotinib, afatinib, osimertinib, and mavelertinib (PF-06747775). Analyses of 28 patients revealed molecular responders had a near complete elimination of ctDNA (>98%) on therapy. Molecular non-responders with limited or no reduction of ctDNA levels experienced a statistically shorter PFS (1.6 vs. 13.7 mos, P22).
Circulating tumor DNA has been detected in urine, and drug induced apoptosis has been modeled within days of TKI treatment in patients with detectable EGFR mutations. In a proof of concept study, we identified an initial spike within the first week of therapy followed by a significant decrease in the number of copies detected from baseline within a week. This work demonstrates that frequent ctDNA sampling may enable early evaluation of patient response or progression (43).
In a separate study, changes in EGFR mutation have been shown to correlate with early clinical response prediction to EGFR TKIs. EGFR-mutated tumors with ctDNA testing at baseline and serially during erlotinib therapy showed a decrease in 95% of cases. The rate of the decrease in ctDNA fraction correlated with radiological response (P50–70%) had longer progression free survival (44).
Recent data from the FLAURA first line osimertinib study of 489 patients showed that the clearance of ctDNA at weeks 3 or 6 was associated with a longer PFS than those patients in whom there was no clearance of ctDNA. Plasma ctDNA analysis of treatment naïve patients with stage III/IV NSCLC were evaluated by ddPCR. EGFR mutation analysis was done at baseline, week 3, and week 6 after EGFR-TKI therapy. Early clearance of plasma EGFR mutation after EGFR-TKI therapy was associated with improved PFS. Patients with detectable baseline plasma ctDNA had shorter PFS than those without detectable EGFR mutation (45). The integration of ctDNA analyses in clinical trials to understand their correlation with clinical outcomes is an ongoing path forward.
Conclusions
There has been a pressing need for a highly sensitive and reliable non-invasive liquid biopsy strategy for screening and resistance monitoring. EGFR testing and tracking through serial ctDNA plasma analyses is being utilized for initial genotyping and resistance monitoring. Plasma assays are being increasingly utilized when tissue is not available. These tests may have high sensitivities and specificities, and several studies are ongoing demonstrating concordance with tissue testing. There has been integration of ctDNA analyses in clinical trials as endpoints to track with response. The development of additional assays will further improve our knowledge of drug resistance, clonal evolution, and combinatorial therapeutic strategies forward.
Acknowledgments
We would like to acknowledge Christian D. Ramirez for assistance in formatting.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Trever G. Bivona) for the series “Mechanisms of Resistance to EGFR-targeted Therapy” published in Journal of Thoracic Disease. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: The series “Mechanisms of Resistance to EGFR-targeted Therapy” was commissioned by the editorial office without any funding or sponsorship. AB has participated on an Advisory board for Astra Zeneca and Pfizer. HH has received research funding from Pfizer, speaker and consultant fees from Astrazeneca, Merck, Takeda, and Bristol Myers Squibb, and has participated on advisory boards with Foundation Medicine and Astrazeneca and Boehringer Ingelheim. AL has no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic Lymphoma Kinase Inhibition in Non–Small-Cell Lung Cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann Oncol 2015;26:1415-21. [Crossref] [PubMed]
- Herbreteau G, Vallée A, Charpentier S, et al. Circulating free tumor DNA in non-small cell lung cancer (NSCLC): clinical application and future perspectives. J Thorac Dis 2019;11:S113-26. [Crossref] [PubMed]
- Saarenheimo J, Eigeliene N, Andersen H, et al. The Value of Liquid Biopsies for Guiding Therapy Decisions in Non-small Cell Lung Cancer. Front Oncol 2019;9:129. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795-801. [Crossref] [PubMed]
- Health CfDaR. Recently-Approved Devices - cobas® EGFR Mutation Test v2 - P150047. 2017. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P150047
- Takano T, Ohe Y, Sakamoto H, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol 2005;23:6829-37. [Crossref] [PubMed]
- Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. [Crossref] [PubMed]
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell-Free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-Small Cell Lung Cancer. Clin Cancer Res 2019;25:4691-700. [Crossref] [PubMed]
- Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014;14:294. [Crossref] [PubMed]
- Kim ST, Jung HY, Sung JS, et al. Can serum be used for analyzing the EGFR mutation status in patients with advanced non-small cell lung cancer? Am J Clin Oncol 2013;36:57-63. [Crossref] [PubMed]
- Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016;7:65364-73. [Crossref] [PubMed]
- Wu YL, Lee JS, Thongprasert S, et al. Intercalated combination of chemotherapy and erlotinib for patients with advanced stage non-small-cell lung cancer (FASTACT-2): a randomised, double-blind trial. Lancet Oncol 2013;14:777-86. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Chaudhuri AA, Binkley MS, Osmundson EC, et al. Predicting Radiotherapy Responses and Treatment Outcomes Through Analysis of Circulating Tumor DNA. Semin Radiat Oncol 2015;25:305-12. [Crossref] [PubMed]
- Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017. [Crossref] [PubMed]
- Phallen J, Leal A, Woodward BD, et al. Early Noninvasive Detection of Response to Targeted Therapy in Non-Small Cell Lung Cancer. Cancer Res 2019;79:1204-13. [Crossref] [PubMed]
- Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385-9. [Crossref] [PubMed]
- @US_FDA. cobas EGFR Mutation Test v2 | FDA. US_FDA. 2019. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/cobas-egfr-mutation-test-v2
- Thierry AR, El Messaoudi S, Gahan PB, et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016;35:347-76. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res 2011;17:1616-22. [Crossref] [PubMed]
- Nygaard AD, Holdgaard PC, Spindler KL, et al. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br J Cancer 2014;110:363-8. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov 2013;3:658-73. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov 2016;6:147-53. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015;26:731-6. [Crossref] [PubMed]
- Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:841-9. [Crossref] [PubMed]
- Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin Cancer Res 2016;22:4837-47. [Crossref] [PubMed]
- Kim TM, Song A, Kim DW, et al. Mechanisms of Acquired Resistance to AZD9291: A Mutation-Selective, Irreversible EGFR Inhibitor. J Thorac Oncol 2015;10:1736-44. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mediates resistance to AZD9291 in advanced non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017;545:446-51. [Crossref] [PubMed]
- Hyun MH, Sung JS, Kang EJ, et al. Quantification of circulating cell-free DNA to predict patient survival in non-small-cell lung cancer. Oncotarget 2017;8:94417-30. [Crossref] [PubMed]
- Lee Y, Park S, Kim WS, et al. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stage EGFR-mutated non-small cell lung cancer. Thorac Cancer 2018;9:1104-10. [Crossref] [PubMed]
- Husain H, Melnikova VO, Kosco K, et al. Monitoring Daily Dynamics of Early Tumor Response to Targeted Therapy by Detecting Circulating Tumor DNA in Urine. Clin Cancer Res 2017;23:4716-23. [Crossref] [PubMed]
- Marchetti A, Palma JF, Felicioni L, et al. Early Prediction of Response to Tyrosine Kinase Inhibitors by Quantification of EGFR Mutations in Plasma of NSCLC Patients. J Thorac Oncol 2015;10:1437-43. [Crossref] [PubMed]
- Zhou C, Imamura F, Cheng Y, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol 2019;37:9020. [Crossref]


