Methylated APC and RASSF1A in multiple specimens contribute to the differential diagnosis of patients with undetermined solitary pulmonary nodules
Introduction
Lung cancer is the leading cause of cancer mortality worldwide. Despite advances in therapeutic options, the overall 5-year survival rate remains less than 15% (1-3). At the time of diagnosis, only one-third of the patients have disease suitable for surgery, the most effective treatment for lung cancer. Early-stage detection is thus the only way to improve its resectability and, likely, its prognosis.
Recent evidence shows that, in addition to genetic changes, epigenetic mechanisms, particularly DNA hypermethylation, are strongly associated with the silencing of several tumor-suppressor genes (TSGs) in lung cancer. Aberrant hymethylation of CpG dinucleotides in the promoter region is a common feature of human cancer and is an important regulatory factor of gene expression (4). Several genes have been well studied, such as MGMT, p16 INK4A, RASSF1A, APC, CHD13, and RARβ (5-8). Many investigators have reported that microsatellite alterations and gene mutations could be identified in the plasma and/or serum DNA of various cancer patients (9-13), which were thought to be released from cancer cells (14-18). Thus circulating tumor derived DNA might be used as a source for tumor detection.
However, the methylation status in different matched samples still remains unclear. The aim of this study was to identify methylated DNA in biopsy tissues, serum and plasma of patients with abnormal findings on their chest radiograph as detected by high-risk lung cancer screening. By comparison with protein markers, it was designed to investigate the role of methylated APC and RASSF1A in the differential diagnosis of solitary pulmonary nodules.
Materials and methods
Subjects
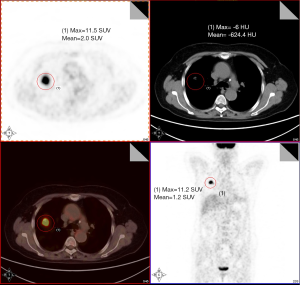
The study population consisted of 89 patients with CT-detected undetermined solitary pulmonary nodules (Figure 1). All the 89 subjects with clinical respiratory symptoms and CT signs suggestive of lung cancer were at high risk of developing cancer. Twenty-three healthy volunteers were also enrolled in the study. The control group matched well with disease group in age, gender, tobacco and so on. Informed consent was obtained according to guidelines from the fields of medical ethics and research ethics. This informed consent has been given based upon a clear appreciation and understanding of the facts, implications, and consequences of blood and tissue detection.
Sample collection
Peripheral blood of all the 89 undetermined solitary pulmonary nodules patients was collected before biopsy. Blood samples were centrifuged at 1,600 g for 10 minutes, and plasma was carefully removed from the ethylenediamine tetraacetic acid (EDTA)-containing tubes, transferred to plain EP tubes. After a second stage centrifugation with 16,000 g for 10 minutes, the supernatant plasma was collected and stored at −70 °C until further processing. Serum samples were also collected after a two-step centrifugation. Plasma and serum samples of 23 healthy blood donors acted as controls. The needle biopsy tissues were immediately stored in liquid nitrogen after removing from patients.
DNA extraction
DNA was extracted from 200 μL plasma or serum samples with internal control by using the BILATEST Viral DNA/RNA Kit (BILATEC, Viernheim, Germany) according to the manufacturer’s recommendations. Tissue DNA was obtained by traditional phenol-chloroform extraction method. DNA samples were stored at −70 °C for subsequent analysis.
Bisulfite modification
Genomic DNA was modified by treatment with sodium bisulfite which converts all non-methylated cytosines to uracil, then to thymidine during the subsequent PCR step. Chemical modification of the DNA was performed using the CpGenomeTM DNA Modification Kit (Chemicon, USA & Canada). Briefly, 1 μg of genomic DNA was denatured by treatment with NaOH and modified by sodium bisulfite. DNA was then bound to a micro-particulate carrier and was desalted by repeated centrifugation and resuspension in 70% ethanol. The conversion to uracil was completed by alkaline desulfonation and desalting was repeated in 90% ethanol. The DNA was finally eluted from the carrier by heating in TE buffer.
Quantitative methylation-specific PCR
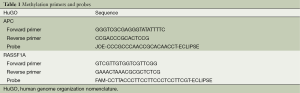
DNA methylation was analyzed by quantitative methylation-specific PCR. All the methylation-specific primes and probes used in this study are designed by Primer Express® Software Version 3.0 and listed in Table 1. DNA extracted from large lung cancer cell line H460 cells was used as RASSF1A and APC methylated or positive control (sequencing confirmed) for the methylation analysis. DNA from healthy volunteers and distilled water were served as an unmethylated or negative control and a blank control. Reaction system content 0.5 μL hot tag, 2.0 μL APC forward primer (5 μm), 2.0 μL APC reverse primer (5 μm), 1.5 μL APC probe (5 μm), 2.0 μL RASSF1A forward primer (5 μm), 2.0 μL RASSF1A reverse primer (5 μm), 1.5 μL RASSF1A probe (5 μm), 10.0 μL 5× buffer, 4.0 μL MgCl2 (25 mm), 2.0 μL dNTPs (10 mm), add ddH2O to 50 μL. The condition was 94 °C for 10 sec, followed by 55 cycles of 95 °C for 5 sec, 60 °C for 34 sec. The standard curve was created by using a series of 1:10 dilutions to calculate the copy numbers of methylated RASSF1A and APC in plasma and serum with concentrations of 150,000, 15,000, 1,500 and 150 copies/mL. The technical triplicates were used in QMSP and at least two positive results could be defined as methylated.
Full table
Statistical analysis
Fisher’s exact test was used to examine the association of different categorical variables. A P value of less than 0.05 was considered to be statistically significant. Spearman’s rank correlation coefficient was used to describe the correlation between two variables. The odds ratios and its 95% confidence intervals (CI) were estimated for the occurrence of lung cancer.
Results
Clinicopathologic diagnosis of patients
Among 89 CT-detected undetermined solitary pulmonary nodules patients, 58 patients were diagnosed with early stage lung cancer (T1a, T1b and T2a staging included) and 31 had benign lung disease according to pathological reports. Twenty-six lung cancer patients had matched biopsy tissue, plasma and serum samples.
Methylation frequency of RASSF1A and APC
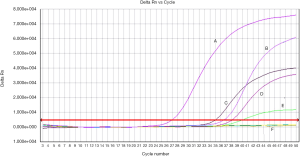
Methylation status of RASSF1A and APC for 89 patients as well as 23 normal samples was examined using QMSP. Representative results of methylation analysis by QMSP were shown in Figure 2, and detailed data regarding the frequency of aberrant methylation were summarized in Table 2.
Full table
For RASSF1A, the methylation rate of three different sample types (plasma, serum and tissue) all showed significant differences among three groups. Significant differences for APC were observed in plasma and tissues. However, the benign disease group showed a relatively lower methylation frequency. No methylation for both of the two genes was found in the healthy group. The difference between benign disease group and healthy group was not significant. Besides, the smoking rates of caner group and benign group were both around 60%.
Association with clinicopathologic features
We analyzed the correlations between methylation status and clinicopathologic variables of the patients. As described in Table 3, no significant difference was found between methylation frequency and age, gender, tobacco or histology. There was an exception for methylation difference with histology of APC. The difference was significant with a P value less than 0.05 (P=0.039). APC methylation seemed to be more common in undifferentiated carcinoma.
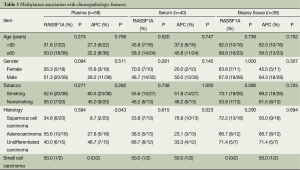
Full table
Powers of RASSF1A and APC methylation assay and NSE and CYFRA 21-1 assay in identifying lung cancer from benign disease
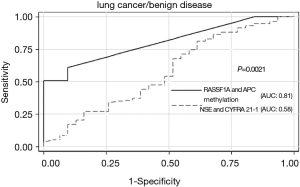
To determine the powers of DNA methylation and protein biomarkers (NSE and CYFRA 21-1) in identifying cancer from benign disease, ROC curves were calculated (Figure 3). RASSF1A and/or APC methylation assay had an area of 0.81 under the ROC curve. By using the optimal cutoff value of 150 copies/mL, we got a sensitivity of 56.9% and a specificity of 90.3%. The AUC of CYFRA 21-1 and NSE assay was 0.58, sensitivity of 48.3%, and specificity of 54.8% (using clinical cutoff value). Consequently, the power of RASSF1A and APC methylation assay was superior to that of NSE and CYFRA 21-1 with a P value of 0.0021.
Combination methylation results
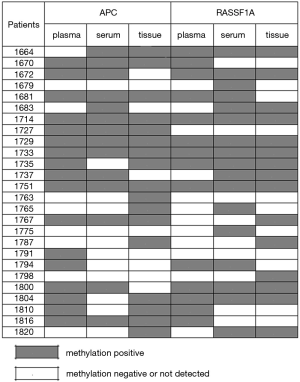
Considering the condition of at least one gene methylation of RASSF1A and APC, we got a much higher methylation frequency list in Table 4. For lung cancer patients, we combined three kinds of samples as well as two genes all together and the methylation rate was 88.5%. Among the 26 early stage lung cancer patients, only three patients showed no methylation for both RASSF1A and APC in three different sample types (Figure 4). The coincidence rate of methylation between plasma and serum of the 26 early stage lung cancer patients was 65.4% for RASSF1A and 73.1% for APC. For RASSF1A and APC, the methylation comparation between plasma and biopsy tissue samples both showed a coincidence rate of 57.7%. The coincidence rate between serum and biopsy tissue samples was 65.4% with RASSF1A and 57.7% with APC. As for RASSF1A, twelve patients’ methylation status of biopsy tissue matched the plasma and serum and the coincidence rate attains 46.2%. It displayed the same as for APC. There were eight patients’ positive methylation results in blood did not match their biopsy tissue results for RASSF1A. And for APC, the number is six. Also, three patients’ positive biopsy tissue methylation results did not match their blood's detection results for RASSF1A. And for APC, there were four cases.

Full table
Sample type comparison
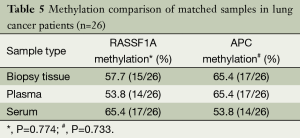
As listed in Table 5, the methylation rates of RASSF1A and APC of paired biopsy tissue, plasma and serum samples were all more than 50%. Data analysis revealed that the differences were not significant.
Full table
Methylation and risk of lung cancer
Figure 5 showed the results of the correlation between number of methylated genes and risk of lung cancer. The diagnostic value of each gene was graphed as a forest plot. Methylation of either RASSF1A or APC in plasma was association with the odds of lung carcinoma. The estimated odds ratio for diagnosis was 15.55 with paired methylated RASSF1A and APC (P=0.000).

Discussion
As of 2008, lung cancer was the leading cause of cancer death in males, as well as the second leading cause of cancer death in females (19). Despite many attempts, the development of a clinically useful protein biomarker for lung cancer has remained elusive. Recently, most studies were focusing on methylation detection of resected cancer tissues in stage II or even later stage lung carcinoma patients. In this research, the study group of 112 subjects comprised of 89 CT-detected undetermined solitary pulmonary nodules patients with no other clinical symptoms and whose tumor size were all less than 5 cm and 23 healthy volunteers. Biopsy was performed under CT conduction to get the histopathologic report which was now being considered the gold standard of diagnosis. As a result, 58 patients were diagnosed with stage I lung cancer (stage T1a, b and T2a included). Undoubtedly, the data in our study could reveal the important role of gene methylation in differential diagnosis of patients with undetermined solitary pulmonary nodules.
Compared to the reported methods and resulted mentioned formerly (20), we got a relative higher methylation frequency. In order to catch the microamount of methylated tumor derived DNAs in peripheral blood firstly we used a good DNA extraction kit and then the QMSP. This definitely explained and confirmed the high methylation frequency we could detect in plasma or serum of stage I lung cancer patients. Considering the serum samples detection performed by Fujiwara et al. and Wang et al. (21,22), the results were all similar to this article (no statistical difference). Compared the results of plasma and serum, the statistical difference was observed in plasma group (P<0.05) but not in serum. At this point, there might be a hypothesis that plasma could be used for methylation detection of early stage lung cancer and further confirm should be designed.
In the current study, samples from benign lung diseases were also investigated for methylation status. Promoter hypermethylation could be detected in two or three patients. One possibility was that some risk factors have been reported to correlate with gene methylation such as smoking status (23). Another hypothesis was that accumulation of DNA methylation might be associated with tumor genesis while some non-cancer diseases could act as a disease process (24). This was also supported by Matsuda that TSGs were inactivated just by extensive CpG methylation (25). Compared with the lung cancer group, the detection level of benign controls is very low. Long-time follow up and further study should be carried out.
One interesting aspect of the present study is that APC methylation was correlated with histopathological type in lung cancer patients. Undifferentiated carcinoma exhibited a much higher methylation frequency than any other type. Different results obtained by different studies conflicted upon this point, especially in the relationship between promoter methylation and clinical features. Stephen’s study revealed that methylation of APC, CCND2, KCNH5 and RUNX was significantly more frequent in adenocarcinoma compared to squamous cell carcinoma (26). And Tomizawa et al. also found a significant methylation difference between adenocarcinoma and squamous cell carcinoma as 39% to 13% (27). The methylation rate of RASSF1A and CDH13 was more frequent in invasive tumors than in noninvasive tumors which suggested its potential important values (28). However, another study group suggested that methylation was independent of clinical features (29). The differences among studies may due to various cancer types, numbers of samples, regions and races as well as methods of extraction, modification and amplification and other factors (30-33). Whether there was correlation between aberrant methylation and lung cancer clinical characteristics needs further analysis to clarify.
Existing protein biomarkers such as CYFRE21-1 and NSE are lack of high sensitivity, specificity and of limited value for early detection (34,35). The data in our research showed that RASSF1A and/or APC methylation assay has a much higher sensitivity of 56.9% and a specificity of 90.3% than CYFRA 21-1 and NSE assay. Several studies have indicated that hypermethylation of CpG islands in gene promoters can occur early in the progression of lung cancer or can be characteristic of premalignant lesion (36,37). Promoter methylation of p16 has been demonstrated from bronchial basal cell hyperplasia to squamous metaplasia to carcinoma in situ with an increasing frequency of 17%, 24% and 50% respectively (38). The CpG island methylator phenotype status of some TSGs displayed differently between NSCLC and paired normal tissues (39). A newly research revealed that methylation of TSGs could be detected in histological normal lymph nodes of lung cancer patients probably confirmed its role as an early event (40). RASSF1A and APC are TSGs in widely research related to carcinogenesis silenced by promoter methylation. Not only our preview studies but also other researchers have all demonstrated the roles of RASSF1A and APC as key TSGs that involved in epigenetic silencing (41-51).
In order to be of value in identification of patients at increased risk of cancer, a marker should be detectable in a biological sample that can be obtained using a minimally invasive procedure. In addition, alterations in the marker should be specific for the relevant premalignant condition or malignancy and should be measurable in patients with early stage cancer or those with premalignant lesions at high risk of progressing to malignancy. Many studies have focused on the detection of genetic and epigenetic abnormalities in exfoliated cells from sputum (52), bronchoalveolar lavage, as well as in the circulating DNA found in plasma or serum. Tumor tissues were good samples for research because abundant DNAs can be acquired. However, obtaining tumor tissue is an invasive procedure and a skill dependent work. Other samples, such as bronchoalveolar lavage, were just applicable on central type lung carcinoma (53). As to sputum, sample collection was not so easy to some patients and the methylation frequency was also very low according to previous studies (54). However, there is still a long way from research to clinical application.
In this study, a few cancer patients did not get positive tissue results when their blood tests showed positive. One possible reason was that the tissue here was obtained just by biopsy not by surgery and the biopsy tissues might not 100% represent cancer tissues because of doctors’ medical skills or some other factors. A few pathological reports of biopsy samples just found necrosis tissues could support this point. So the detection of resected tumor tissues could reveal the real methylation status. There remained some other cases whose biopsy tissues were positive methylation but plasma and serum both got negative results. This was probably because all the enrolled cases were early stage lung cancer and the amount of cancer-related methylated TSGs in blood might mainly depend on the tumor size and cancer stage. However, the concordance of methylation in peripheral blood and tumor tissue might be the most valuable evidence in the diagnosis of early stage lung cancer.
In the present study, we used multiple samples including plasma, serum and biopsy tumor tissues not only to improve methylation rate but also aimed to find out a much more suitable sample for DNA methylation detection. Firstly, our results of combined methylation rate of lung cancer patients had been highly improved to 88%. Secondly, methylation comparison of paired samples in lung cancer patients revealed that the methylation rates of the three different sample types were all around 60% and there was no significant difference among plasma, serum and biopsy tumor tissues. So we chose plasma as specimen with advantages of easy collection, minimal invasion and repetitive test.
Conclusions
In conclusion, compared with cancer-free controls, detection of promoter hypermethylation on these two genes was much more frequent in early stage lung cancer patients. The RASSF1A and APC methylation assay could be very useful for the differential diagnosis of patients with undetermined solitary pulmonary nodules. These molecular changes may be a valuable biomarker for early detection and risk assessment in high-risk populations.
Acknowledgements
Funding: This work was supported by grants from Key Laboratory for Laboratory Medicine of Jiangsu Province of China (No. XK201114), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the National Natural Science Foundation of China (81101322, 81201359 and 81371894) and the Natural Science Foundation of Jiangsu Province (BK2009440).
Disclosure: The authors declare no conflict of interest.
References
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [PubMed]
- Yang L, Parkin DM, Li LD, et al. Estimation and projection of the national profile of cancer mortality in China: 1991-2005. Br J Cancer 2004;90:2157-66. [PubMed]
- Pastorino U. Lung cancer screening. Br J Cancer 2010;102:1681-6. [PubMed]
- Esteller M. Epigenetics in cancer. N Engl J Med 2008;358:1148-59. [PubMed]
- Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer 2004;4:707-17. [PubMed]
- Pan S, Zhang L, Gao L, et al. The property of methylated APC gene promotor and its influence on lung cancer cell line. Biomed Pharmacother 2009;63:463-8. [PubMed]
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [PubMed]
- Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. RARβ2 gene methylation level in the circulating DNA from blood of patients with lung cancer. Eur J Cancer Prev 2011;20:453-5. [PubMed]
- Chen XQ, Stroun M, Magnenat JL, et al. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med 1996;2:1033-5. [PubMed]
- Yazici H, Terry MB, Cho YH, et al. Aberrant methylation of RASSF1A in plasma DNA before breast cancer diagnosis in the Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev 2009;18:2723-5. [PubMed]
- Hibi K, Robinson CR, Booker S, et al. Molecular detection of genetic alterations in the serum of colorectal cancer patients. Cancer Res 1998;58:1405-7. [PubMed]
- Cho YH, Yazici H, Wu HC, et al. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res 2010;30:2489-96. [PubMed]
- Mulcahy HE, Lyautey J, Lederrey C, et al. A prospective study of K-ras mutations in the plasma of pancreatic cancer patients. Clin Cancer Res 1998;4:271-5. [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Van der Auwera I, Elst HJ, Van Laere SJ, et al. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br J Cancer 2009;100:1277-86. [PubMed]
- Matuschek C, Bölke E, Lammering G, et al. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res 2010;15:277-86. [PubMed]
- Chen Z, Fadiel A, Naftolin F, et al. Circulation DNA: biological implications for cancer metastasis and immunology. Med Hypotheses 2005;65:956-61. [PubMed]
- Skvortsova TE, Rykova EY, Tamkovich SN, et al. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br J Cancer 2006;94:1492-5. [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [PubMed]
- Tomizawa Y, Kohno T, Kondo H, et al. Clinicopathological significance of epigenetic inactivation of RASSF1A at 3p21.3 in stage I lung adenocarcinoma. Clin Cancer Res 2002;8:2362-8. [PubMed]
- Fujiwara K, Fujimoto N, Tabata M, et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res 2005;11:1219-25. [PubMed]
- Wang Y, Yu Z, Wang T, et al. Identification of epigenetic aberrant promoter methylation of RASSF1A in serum DNA and its clinicopathological significance in lung cancer. Lung Cancer 2007;56:289-94. [PubMed]
- Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res 2006;66:3338-44. [PubMed]
- Oue N, Mitani Y, Motoshita J, et al. Accumulation of DNA methylation is associated with tumor stage in gastric cancer. Cancer 2006;106:1250-9. [PubMed]
- Matsuda Y, Ichida T, Matsuzawa J, et al. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology 1999;116:394-400. [PubMed]
- Hawes SE, Stern JE, Feng Q, et al. DNA hypermethylation of tumors from non-small cell lung cancer (NSCLC) patients is associated with gender and histologic type. Lung Cancer 2010;69:172-9. [PubMed]
- Tomizawa Y, Iijima H, Nomoto T, et al. Clinicopathological significance of aberrant methylation of RARbeta2 at 3p24, RASSF1A at 3p21.3, and FHIT at 3p14.2 in patients with non-small cell lung cancer. Lung Cancer 2004;46:305-12. [PubMed]
- Kubo T, Yamamoto H, Ichimura K, et al. DNA methylation in small lung adenocarcinoma with bronchioloalveolar carcinoma components. Lung Cancer 2009;65:328-32. [PubMed]
- Ito M, Ito G, Kondo M, et al. Frequent inactivation of RASSF1A, BLU, and SEMA3B on 3p21.3 by promoter hypermethylation and allele loss in non-small cell lung cancer. Cancer Lett 2005;225:131-9. [PubMed]
- Xu X, Gammon MD, Zhang Y, et al. Gene promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat 2010;121:685-92. [PubMed]
- Lind GE, Danielsen SA, Ahlquist T, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer 2011;10:85. [PubMed]
- Azuara D, Rodriguez-Moranta F, de Oca J, et al. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clin Colorectal Cancer 2010;9:168-76. [PubMed]
- Tänzer M, Balluff B, Distler J, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One 2010;5:e9061. [PubMed]
- Keller T, Bitterlich N, Hilfenhaus S, et al. Tumour markers in the diagnosis of bronchial carcinoma: new options using fuzzy logic-based tumour marker profiles. J Cancer Res Clin Oncol 1998;124:565-74. [PubMed]
- Watine J. Prognostic evaluation of primary non-small cell lung carcinoma patients using biological fluid variables. A systematic review. Scand J Clin Lab Invest 2000;60:259-73. [PubMed]
- Worthley DL, Whitehall VL, Buttenshaw RL, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene 2010;29:1653-62. [PubMed]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003;349:2042-54. [PubMed]
- Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A 1998;95:11891-6. [PubMed]
- Liu Z, Zhao J, Chen XF, et al. CpG island methylator phenotype involving tumor suppressor genes located on chromosome 3p in non-small cell lung cancer. Lung Cancer 2008;62:15-22. [PubMed]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med 2008;358:1118-28. [PubMed]
- Ng EK, Leung CP, Shin VY, et al. Quantitative analysis and diagnostic significance of methylated SLC19A3 DNA in the plasma of breast and gastric cancer patients. PLoS One 2011;6:e22233. [PubMed]
- Cho YH, Shen J, Gammon MD, et al. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat 2012;131:197-205. [PubMed]
- Ahmed IA, Pusch CM, Hamed T, et al. Epigenetic alterations by methylation of RASSF1A and DAPK1 promoter sequences in mammary carcinoma detected in extracellular tumor DNA. Cancer Genet Cytogenet 2010;199:96-100. [PubMed]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107-16. [PubMed]
- Hill VK, Hesson LB, Dansranjavin T, et al. Identification of 5 novel genes methylated in breast and other epithelial cancers. Mol Cancer 2010;9:51. [PubMed]
- Dietrich D, Krispin M, Dietrich J, et al. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC Cancer 2010;10:247. [PubMed]
- Pan SY, Xie EF, Shu YQ, et al. Methylation quantification of adenomatous polyposis coli (APC) gene promoter in plasma of lung cancer patients. Ai Zheng 2009;28:384-9. [PubMed]
- Zhang LX, Pan SY, Chen D, et al. Effect of adenomatous polyposis coli(APC) promoter methylation on gene transcription in lung cancer cell lines. Ai Zheng 2007;26:576-80. [PubMed]
- Fleischhacker M, Dietrich D, Liebenberg V, et al. The role of DNA methylation as biomarkers in the clinical management of lung cancer. Expert Rev Respir Med 2013;7:363-83. [PubMed]
- Liu WJ, Tan XH, Guo BP, et al. Associations between RASSF1A promoter methylation and NSCLC: a meta-analysis of published data. Asian Pac J Cancer Prev 2013;14:3719-24. [PubMed]
- Sheikhnejad R, Zohri M, Shadmehr MB, et al. Detection of aberrant methylation of 10 genes in genomic DNA of lung tumors. Ann Oncol 2013;24:2705-6. [PubMed]
- van der Drift MA, Prinsen CF, Hol BE, et al. Can free DNA be detected in sputum of lung cancer patients? Lung Cancer 2008;61:385-90. [PubMed]
- de Fraipont F, Moro-Sibilot D, Michelland S, et al. Promoter methylation of genes in bronchial lavages: a marker for early diagnosis of primary and relapsing non-small cell lung cancer? Lung Cancer 2005;50:199-209. [PubMed]
- Cirincione R, Lintas C, Conte D, et al. Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study. Int J Cancer 2006;118:1248-53. [PubMed]