Distribution of pathogenic bacteria in lower respiratory tract infection in lung cancer patients after chemotherapy and analysis of integron resistance genes in respiratory tract isolates of uninfected patients
Introduction
In recent years, the incidence and mortality of lung cancer have become the highest among malignant tumors (1-3). Early stage lung cancer patients always have no symptoms, so most lung cancer patients are in the middle or late stage when detected and lost the opportunity of surgical treatment, chemotherapy is the classic treatment for these patients. In recent years, immunotherapy is rising gradually and becomes a new choice (4-6). The immunity of patients with lung cancer, especially in patients after chemotherapy, is very low, so the patients of lung cancer treated with chemotherapy are more easily to suffer pulmonary infection (7). Meanwhile, pulmonary infection is one of the causes of death in patients with lung cancer undergoing chemotherapy (8). Therefore, it is necessary to monitor the drug sensitivity of pathogens in patients with lung cancer complicated with pulmonary infection after chemotherapy. Previous studies on drug resistance of bacteria mainly focused on gene mutation of bacterial chromosome, but this kind of bacterial drug resistance caused by gene mutation cannot be transmitted. Studies have found that the reason for most bacteria to develop drug resistance is to obtain exogenous drug resistance genes (9). It has been shown that drug resistance genes can be transmitted horizontally through conjugated plasmids, transposons and integrated phages (10-12). Stokes and colleagues first reported integron, which was related to the horizontal transmission of drug resistant genes (13-15). Integron is a kind of movable gene element, carrying site-specific recombination system components, which can integrate many drug-resistant gene cassettes together to form multiple drug resistance. It plays an important role in the spread of drug-resistance of bacteria, especially gram-negative bacteria (16). A lot of studies have researched the isolated strains of infected patients; few of them pay attention to the integron carried by the respiratory tract strains of uninfected patients. Many kinds of hospital strains can be planted in the respiratory tract of patients undergoing chemotherapy, who have been hospitalized for several times. In order to better monitor and control the spread of bacterial resistance, the integron carried by the isolated bacteria of respiratory tract samples of lung cancer patients without pulmonary infection after chemotherapy is detected. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-928).
Methods
Patient collection
We collected 400 patients who were hospitalized in Fuyang People’s Hospital from July 2017 to July 2019. All patients were diagnosed with lung cancer and received chemotherapy. Pulmonary infection was diagnosed according to the relevant diagnostic criteria of respiratory branch of Chinese Medical Association. The patients present COPD are excluded. Among the patients, 261 were male and 139 were female. The average age was (56±9.5) years. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided written consent. The study was approved by the Ethics Committee of Fuyang People’s Hospital.
Sputum collection
Patients were rinsed with normal saline in the morning, and the deep cough sputum was kept in a sterile sputum box for examination within 1 hour. The strain was identified by the bacterial identification system of Biomerieux of France. The K-B method recommended by WHO was used to detect the sensitivity of the strains to antibiotics. The quality control strains were Klebsiella pneumoniae ATCC700603, Escherichia coli ATCC25922, Staphylococcus aureus ATCC25923, Streptococcus pneumoniae ATCC29213, Pseudomonas aeruginosa ATCC27853 and Candida albicans ATCC10231. The standard reference strain and the experimental strain were determined in parallel during the experiment.
Preparation of total bacterial DNA
The bacterial DNA was extracted according to the operating instructions of the bacterial DNA extraction kit and analyzed by electrophoresis.
Integron detection and classification
To detect integron, the class I, class II and class III integrase genes in the 5' conserved region of integron were amplified simultaneously by annexation primers, this process also known as integrase gene polymerase chain reaction (PCR) (17). The sequence of the upstream primer hep35 was 5'-tgcgggtyarargatbtkgattt-3', and the sequence of the downstream primer hep36 was 5'-carcacatgcgtrtaat-3'. The expected amplification fragment is 491 bp, the amplification system contained 25 µL premix Taq, 2 µL template, 1 µL upstream and downstream primers each, 21 µL sterile distilled water, the total circulation system was 50 µL. The amplified products of integrase PCR were analyzed by restriction endonuclease Hinf I digestion. 2 µL 10×H Buffer, 1 µL Hinf I enzyme and 8 µL PCR product were bathed in water at 37 degrees centigrade for 4 hours together. The enzyme products were analyzed by agarose 2% gel electrophoresis. According to the results of electrophoretic, class I integron was defined as a 491 bp fragment of the amplified product of integrase PCR after digestion.
Statistical analysis
SPSS 17.0 was used to perform statistical analysis. We analyzed the detection rate of population pathogen and drug resistance rate of different bacteria.
Results
Patient clinical data
We collected 400 patients who were hospitalized in Fuyang People’s Hospital from July 2017 to July 2019. Among the patients, 261 were male and 139 were female. The average age was (56±9.5) years. Twenty-five of the patients had high blood pressure and the blood pressure was well controlled. None of them had mellitus diabetes or some other co-morbidities.
Distribution of pathogenic bacteria in respiratory tract of patients with lung cancer after chemotherapy
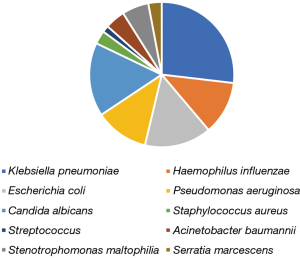
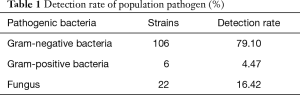
One hundred and thirty-four strains were identified in 400 patients after chemotherapy. There were 120 patients with pulmonary infection, 114 strains were cultured. Among 280 patients without pulmonary infection, 20 strains were cultured; all of them were Klebsiella pneumoniae. Among the 134 cultured strains, the detection rate of gram-negative bacteria was 79.10%, and the top four were Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Haemophilus influenzae. The detection rate of gram-positive bacteria was 4.47%, mainly were Staphylococcus aureus and Streptococcus. Twenty-two strains of fungi were detected, all of them were Candida albicans, and the detection rate was 16.42% (Table 1, Figure 1).
Full table
Drug resistance rate of gram-negative bacilli
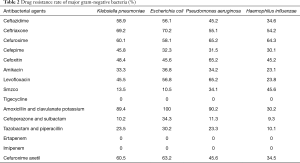
As shown in Table 2, gram negative bacteria (106 strains) had high resistance to penicillin and cephalosporins, and high sensitivity to carbapenems, piperacillin tazobactam and cefoperazone sulbactam.
Full table
Drug resistance rate of gram-positive bacteria
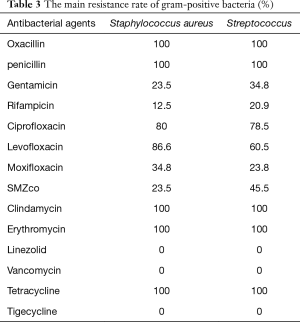
Gram positive bacteria (6 strains) had high resistance to penicillin, macrocyclic lipids and clindamycin, and were sensitive to linezolid, vancomycin and rifampicin (Table 3).
Full table
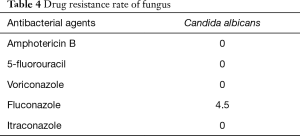
Drug resistance rate of fungi
Table 4 showed that all fungi (22 strains) were Candida albicans, which were sensitive to common antifungal drugs.
Full table
Integron detection and classification
Twenty strains were cultured in 280 patients without pulmonary infection, which were all Klebsiella pneumoniae, among them, 2 strains were integron positive, all of which were class I integron.
Discussion
Chemotherapy is one of the most important treatment methods of advanced lung cancer patients (4-6). However, after chemotherapy, the white cell count of some patients will decrease, the hospitalization time will be extended, and not only the cancer cells but also the normal immune cells will be killed at the same time, thus the lung cancer patients are prone to suffer pulmonary infection after chemotherapy (18). In turn, it is necessary to research the pathogen distribution and drug resistance of pulmonary infection in patients with lung cancer after chemotherapy, which is helpful to guide the clinical rational use of various antibacterial drugs to effectively control infection and eventually reduce the mortality of patients.
In this study, 134 strains of pathogenic bacteria were isolated from the respiratory secretion of 400 patients with lung cancer after chemotherapy. Among them, 106 strains were gram-negative bacteria (79.10%), 22 strains were fungi (16.42%) and 6 strains were gram-positive cocci (4.47%). The results displayed that the pathogens of lung cancer patients with pulmonary infection after chemotherapy were quite different from those of community acquired pneumonia. Studies have shown that the most common pathogens of community acquired pneumonia are Streptococcus pneumoniae, Haemophilus influenzae and atypical pathogens (19-21). It’s better to choose the antibiotics for gram-negative bacilli before the drug sensitivity results come out for lung cancer patients with pulmonary infection after chemotherapy. Compared with other related reports, the detection rate of fungi is higher, while that of gram-positive cocci is lower. The following factors may be related to as followed; first of all, chemotherapy can decrease the number of leukocyte and granulocyte, which will reduce the immunity of patients, so the patients have high risk to get fungal infection. Besides, in the long-term and repeated infection of lung cancer patients, broad-spectrum antibiotics will be used in clinical, which results in the long-term use of antibiotics and increases the possibility of fungal infection. Fungal infection is one of the causes of death in patients with low immunity. It has been found that fungi could produce drug resistance through many mechanisms. There are few drugs to choose in clinical in the treatment of fungal infection, and the fungi are prone to get cross resistance to antifungal drugs, which makes the treatment more difficult in lung cancer patients after chemotherapy (22,23).
In the research, we found that among the 106 strains, the main gram-negative bacteria were Klebsiella pneumoniae, Pseudomonas aeruginosa, Escherichia coli and Haemophilus influenzae. They were highly resistant to penicillin and cephalosporins, and sensitive to carbapenems, piperacillin tazobactam and cefoperazone sulbactam, suggesting that the above three drugs can be used as the first choice for the treatment of pulmonary infection of lung cancer patients after chemotherapy. The result showed that the gram-positive bacteria were Staphylococcus aureus and Streptococcus pneumoniae, and the detection rate of them was relatively low. This result was considered to be related to the distribution of local pathogenic bacteria and the selection and application of antibiotics.
From Table 2, we could see that the strains were highly resistant to penicillin and cephalosporin, and no strains resistant to linezolid and vancomycin were found. Thus, this kind of antibiotics is the first choice to treat multi-resistant Staphylococcus aureus and Streptococcus. We found that the detection rate of fungi was very high, which might be related to the low immunity of lung cancer patients after chemotherapy. It could be seen from Table 3 that, the fungi were sensitive to common antifungal drugs, such as fluconazole, voriconazole and itraconazole, so the common antifungal drugs could be selected to treat fungal infection in clinical.
In recent years, the detection rate of carbapenem resistant Enterobacteriaceae (CRE) is increasing. The treatment of infection caused by CRE is limited, leading to the high mortality (24,25). We found that 20 strains were isolated from respiratory tract of patients without pulmonary infection, and all of them were Klebsiella pneumoniae. Klebsiella pneumoniae, belonging to Enterobacteriaceae, is the most important conditional pathogen in iatrogenic infection except Escherichia coli. Severe or elderly patients are more susceptible to Klebsiella pneumoniae (26). With the wide use of antibiotics, Klebsiella pneumoniae resistant strains continue to increase. The monitoring and the mechanisms of drug resistance of Klebsiella pneumoniae has become the focus of international medicine (27-29).
Some studies have shown that the drug resistance of clinical pathogens, environmental bacteria, animals and normal human flora is increasing with the extensive and excessive use of antibiotics (30). Integron plays a pivotal role in the acquisition and transmission of drug resistance genes. The resistance rate of integron positive strains is significantly higher than that of integron negative strains (13). Therefore, the detection of integron in uninfected patients is conducive to monitoring the spread of drug-resistant genes. Four kinds of integron have been identified, and most clinical isolates bacilli have class I integron (31). So far, most integron types were found in Enterobacteriaceae, and the resistant gene box was the most complex. The main integron types were class 1, class 2 and class 3. Among them, class 1 and class 2 integron were reported more, and class 3 integron was only reported in a case (32,33). A lot of studies focused on the integron carried by infected patients, all class 1 integrons were found in clinical isolated strains. But there were many researchers detected integron in the environment out of the hospital (34-37). We found that, 20 Klebsiella pneumoniae strains were integron positive, all of which were class I integron. The detection of integron in respiratory tract samples of lung cancer patients without pulmonary infection after chemotherapy is of great significance to detect the spread of drug resistance.
Conclusions
After chemotherapy for lung cancer patients, the bacteria isolated from respiratory tract were mainly gram-negative bacteria, with high resistance rate to common penicillin and cephalosporins. The detection rate of gram-positive bacteria was low, and the bacteria were highly resistant to penicillin. Fungi detection rate was high, and sensitive to common antifungal drugs. The clinical isolated strains carried class I integron, which can transmit resistant genes. Therefore, to guide the rational use of antibiotics, the drug sensitivity monitoring should be carried out in time for lung cancer patients with suspected pulmonary infection after chemotherapy. Integron positive bacteria has low toxicity and can survive in the human body for a long time, it can provide the possibility for the epidemic of infectious diseases once conditions are suitable. Therefore, for uninfected patients, it is necessary to detect the spread of drug resistance through the detection of integron.
Acknowledgments
Funding: This study was supported in part by a grant from National Natural Science Foundation of China (81802255), Shanghai Pujiang Program (17PJD036) and a grant from Shanghai Municipal Commission of Health and Family Planning Program (20174Y0131), National Key Research & Development Project (2016YFC0902300), Major Disease Clinical Skills Enhancement Program of Three Year Action Plan for Promoting Clinical Skills and Clinical Innovation in Municipal Hospitals, Shanghai Shen Kang Hospital Development Center Clinical Research Plan of SHDC (16CR1001A), “Dream Tutor” Outstanding Young Talents Program (fkyq1901), Key Disciplines of Shanghai Pulmonary Hospital (2017ZZ02012), Grant of Shanghai Science and Technology Commission (16JC1405900).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-928
Data Sharing Statement: available at http://dx.doi.org/10.21037/jtd-20-928
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-928). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fuyang People’s Hospital. All participants were competent to provide their consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Didkowska J, Wojciechowska U, Mańczuk M, et al. Lung cancer epidemiology: contemporary and future challenges worldwide. Ann Transl Med 2016;4:150. [Crossref] [PubMed]
- Lin CC. Challenges of the phase I drug development in non-small cell lung cancer. Chin Clin Oncol 2019;8:25. [Crossref] [PubMed]
- Watanabe K, Shinkai M, Tei Y, et al. Chemotherapy in non-small cell lung cancer patients receiving oxygen therapy. Oncol Res Treat 2016;39:587-90. [Crossref] [PubMed]
- Rashdan S, Gerber DE. Immunotherapy for non-small cell lung cancer: from clinical trials to real-world practice. Transl Lung Cancer Res 2019;8:202-7. [Crossref] [PubMed]
- Khandekar MJ, Jain R. Smooth sailing for immunotherapy for unresectable stage III non-small cell lung cancer: the PACIFIC study. Transl Cancer Res 2018;7:S16-20. [Crossref] [PubMed]
- Udo EE, Al-Sweih N, John P, et al. Antibiotic resistance of enterococci isolated at a teaching hospital in Kuwait. Diagn Microbiol Infect Dis 2002;43:233-8. [Crossref] [PubMed]
- Humphrey LL, Deffebach M, Pappas M, et al. Screening for lung cancer with low-dose computed tomography: a systematic review to update the US Preventive services task force recommendation. Ann Intern Med 2013;159:411-20. [Crossref] [PubMed]
- Giakkoupi P, Tzouvelekis LS, Tsakris A, et al. IBC-1, a Novel Integron-Associated Class A Beta-Lactamase With Extended-Spectrum Properties Produced by an Enterobacter Cloacae Clinical Strain. Antimicrob Agents Chemother 2000;44:2247-53. [Crossref] [PubMed]
- Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994;264:375-82. [Crossref] [PubMed]
- Waine DJ, Honeybourne D, Smith EG, et al. Association between hypernutator phenotype, clinical variables, mucoid phenotype and antimicrobial resistance in Pseudomonas aeruginosa. J Clin Microbiol 2008;46:3491-93. [Crossref] [PubMed]
- Iyer A, Barbour E, Azhar E, et al. Transposable elements in Eschcherichia coli antimicrobial resistance. Advanc Biosci Biotechnol 2013;4:415-23. [Crossref]
- Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 1989;3:1669-83. [Crossref] [PubMed]
- Swedberg G. Organization of two sulfonamide resistance genes on plasmids of gram-negative bacteria. Antimicrob Agents Chemother 1987;31:306-11. [Crossref] [PubMed]
- Sundström L, Vinayagamoorthy T, Sköld O. Novel type of plasmid-borne resistance to trimethoprim. Antimicrob Agents Chemother 1987;31:60-66. [Crossref] [PubMed]
- Rowe-Magnus DA, Guerout AM, Mazel D. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol Microbiol 2002;43:1657-69. [Crossref] [PubMed]
- Liu H, Wang H, Huang M, et al. Analysis of antimicrobial resistance and class 1 integrons among strains from upper respiratory tract of healthy adults. J Thorac Dis 2013;5:149-55. [PubMed]
- Aliberti S, Amir A, Peyrani P, et al. Incidence, etiology, timing, and risk factors for clinical failure in hospitalized patients with community-acquired pneumonia. Chest 2008;134:955-62. [Crossref] [PubMed]
- Welte T, Torres A, Nathwani D. Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax 2012;67:71-9. [Crossref] [PubMed]
- Drijkoningen JJ, Rohde GG. Pneumococcal infection in adults: burden of disease. Clin Microbiol Infect 2014;20:45-51. [Crossref] [PubMed]
- Song JH, Thamlikitkul V, Hsueh PR. Clinical and economic burden of community-acquired pneumonia amongst adults in the Asia-Pacific region. Int J Antimicrob Agents 2011;38:108-17. [PubMed]
- Mitchell KF, Taff HT, Cuevas MA, et al. Role of matrix ß-1,3 glucan in antifungal resistance of non-albicans Candida biofilms. Antimicrob Agents Chemother 2013;57:1918-20. [Crossref] [PubMed]
- de Silva WJ, Gonçalves LM, Seneviratne J. Exopolysaccharide matrix of developed Candida albicans biofilms after exposure to antifungal agents. Braz Dent J 2012;23:716-22. [Crossref] [PubMed]
- Wang Q, Zhang Y, Yao X, et al. Risk factors and clinical outcomes for carbapenem-resistant Enterobacteriaceae nosocomial infections. Eur J Clin Microbiol Infect Dis 2016;35:1679-89. [Crossref] [PubMed]
- Goodman KE, Simner PJ, Tamma PD, et al. Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev Anti Infect Ther 2016;14:95-108. [Crossref] [PubMed]
- Ye M, Tu J, Bi Y, et al. Clinical and genomic analysis of liver abscess causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol 2016;6:165. [Crossref] [PubMed]
- Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol 2018;45:131-9. [Crossref] [PubMed]
- Leavitt A, Chmelnitsky I, Ofek I, et al. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother 2010;65:243-48. [Crossref] [PubMed]
- Bassetti M, Righi E, Carnelutti A, et al. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Expert Rev Anti Infect Ther 2018;16:749-61. [Crossref] [PubMed]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 2010;74:417-33. [Crossref] [PubMed]
- Meng J, Zhao S, Doyle MP. Virulence Genes of Shiga Toxin-producing Escherichia Coli Isolated from Food, Cattle and Humans. Int J Food Microbiol 1998;45:229-35. [Crossref] [PubMed]
- Collis CM, Kim MJ, Partridge SR, et al. Characterization of the class 3 integron and the site-specific recombination system it determines. J Bacteriol 2002;184:3017-26. [Crossref] [PubMed]
- Correia M, Boavida F, Grosso F, et al. Mplecular characterization of a new class 3 integron in Klebsiella pneumonia. Antimicrob Agents Chemother 2003;47:2838-43. [Crossref] [PubMed]
- Lapara TM, Burch TR, McNamara PJ, et al. Tertiary-Treated Municipal Wastewata is a Significant Point-Source of Antibiotic Resistance Gene into Duluth-Superior Harbor. Environ Sci Techm 2011;10:1021-48.
- Su HC, Ying GG, Tao R, et al. Occurrence of antibiotic resistance and characterization of resistance gene and integrons in Enterobacteriaceae isolated from integrated fish farms in sacth China. Environ Monit 2011;10:1039-63.
- Karczmarczyk M, Walsh C, Slowey R, et al. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. Appl Environ Microbiol 2011;77:7121-7. [Crossref] [PubMed]
- Kumar A, Chakraborti S, Joshi P, et al. A multiple antibiotic and serum resistant oligotrophic strain, Klebsiella pneumoniae MB45 having novel dfrA30, is sensitive to ZnO QDs. Ann Clin Microbiol Antimicrob 2011;10:19. [Crossref] [PubMed]