Expert consensus-based laboratory testing of SARS-CoV-2
At the end of December 2019, Wuhan reported a number of cases of unidentified pneumonia whose pathogen was later identified as a novel coronavirus, by DNA sequencing. As of 8 a.m. on May 6, 2020, novel coronavirus infections have been reported in 214 countries and regions worldwide, posing a huge threat to global public health. China has taken a series of prompt measures of prevention and control while providing medical services, which effectively contained the epidemic domestically. Rapid and accurate laboratory testing plays a crucial role in the etiological diagnosis of COVID-19. In order to improve the understanding of laboratory testing methods for COVID-19 and summarize the cumulative knowledge on the subject, the Guangdong Provincial Department of Science and Technology organized experts in the field of clinical virology diagnosis and treatment (including virology, clinical laboratory, clinical medicine, and preventive medicine), summarized the research progress and experience of novel coronavirus laboratory testing technologies, both locally and abroad, and developed a consensus-based report for novel coronavirus testing. This report covers the aspects of etiology, nucleic acid testing, serological screening, biosafety, and containment in relation to the novel coronavirus and suggests some possible problems that may be encountered in actual testing.
Etiology
The novel coronavirus belongs to the subfamily Sarbecovirus β coronavirus. It is a positive-sense, single-stranded RNA virus and often a pleomorphic enveloped virus. The viral particle is round or ovular with a diameter of 60–140 nm. The novel coronavirus genome encodes four major types of structural proteins, including spike protein (S), nucleocapsid protein (N), membrane protein (M), and envelope protein (E). Its gene sequence is similar to that of the SARS-CoV gene sequence, both of which most likely originated from bats. The homology of the two S proteins in the primary amino acid sequence is 87.2%, and the similarity between the smallest receptor-binding domains is 83.9%, but there are apparent differences in gene characteristics (1). The novel coronavirus S protein mediates receptor binding and membrane fusion, and its S1B domain is recognized by angiotensin converting enzyme (ACE2) receptors on the host cell surface, allowing it to enter the cell. After fusion of the viral envelope with the host cell membrane, the viral genome replicates and transcribes within the cytosol of the host cell. Viral structure proteins and newly synthesized RNA genomes are assembled and modified in the endoplasmic reticulum and Golgi apparatus. Then, new progeny virus particles bud into the endoplasmic reticulum/Golgi intermediate lumen (ERGIC) and are released through vesicles (2). The affinity of the novel coronavirus S protein for ACE2 is about 10 to 20 times higher than that of SARS-CoV, which may accelerate the spread of the novel coronavirus among humans (3). On February 11, 2020, the Coronavirus Research Group of the International Committee of Taxonomy of Viruses (ICTV) recommended that the novel coronavirus be named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and WHO named the pneumonia caused by the novel coronavirus COVID-19.
SARS-CoV-2 is sensitive to ultraviolet light and heat and can usually be inactivated by exposure to 56 °C heat for 30 min. It is an enveloped virus that is sensitive to most organic solvents that affect the membrane, including ether, 75% ethanol, chlorine-containing disinfectants, peroxyacetic acid, chloroform, and other lipid solvents, but chlorhexidine cannot effectively inactivate the virus (4). SARS-CoV-2 is reportedly stable in aerosols and on surfaces for several hours to several days (5).
Laboratory diagnostic methods
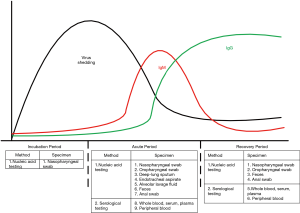
Clinical diagnosis of COVID-19 can be made based on a clear history of epidemiological exposure, systemic symptoms, and chest tomography; however, laboratory testing is important for etiological diagnosis. Laboratory diagnosis involves viral nucleic acid assays, serological testing, and general laboratory analyses. Specimens include nasopharyngeal swabs, oropharyngeal swabs, sputum, other lower respiratory secretions, blood, feces, and saliva. Due to the dynamic changes in viral load and serological markers during the development of the disease, adoption of the appropriate laboratory tests at a specific stage is crucial for clinicians to confirm the diagnosis of COVID-19 (Figure 1).
General testing
Routine blood tests usually show normal or decreased white blood cell counts, decreased lymphocyte counts (progressive lymphopenia in severe cases), normal or elevated C-reactive protein (CRP), and normal procalcitonin (ProPCT) in most cases. Severe cases may exhibit elevated alanine aminotransferase, aspartate aminotransferase, creatine kinase, lactate dehydrogenase, myoglobin, and D-dimer levels. Inflammatory cytokines such as interleukin 6 (IL-6) and IL-10 are often found to be increased in severe and critical patients (6).
Nucleic acid testing
A variety of molecular diagnostic techniques have been applied to SARS-CoV-2 nucleic acid testing, including real-time fluorescent PCR, gene sequencing, isothermal amplification, and POCT molecular diagnostic systems (see Table 1). Real-time fluorescence PCR and gene sequencing are included in the diagnosis and treatment of COVID-19 by the National Health Commission of the PRC in order to provide important etiological evidence for the diagnosis of COVID-19. Laboratories may choose an appropriate diagnostic approach according to the specimens tested along with the sample quantities and their conditions. Currently, the most commonly used technique is real-time fluorescence PCR, which has been elaborated in the following section.
Full table
Reference for reagent selection
Reagents with “internal standard” should be selected. Internal standard reagents detect the transcription of human genes such as RNase P, in order to monitor whether enough human cells were collected in the specimen.
In order to improve the sensitivity and specificity of molecular assays, it is recommended to include at least two loci of SARS-CoV-2 genes (open reading frame 1a/b, nucleocapsid protein N, or envelope protein E) (7) as well as reagents with large loading amounts of nucleic acid templates and large amplification reaction systems.
Since the amplification reaction efficiency may be different for different target genes, it is recommended to use a test kit for a single target to re-test the samples in question.
Pretreatment of specimens
Oropharyngeal swabs and nasopharyngeal swabs can be used directly for nucleic acid extraction. If the personal protective conditions of a laboratory are not sufficient, virus inactivation should be added (7), but it may reduce the sensitivity of nucleic acid testing to some extent (8).
Anticoagulated blood can be centrifuged at 1,500 ×g for 10 min, and the plasma may be used for nucleic acid extraction.
Alveolar lavage fluid and feces should be vortexed before the virus is inactivated.
The sputum needs to be liquefied before nucleic acid extraction. If proteinase K is pre-added, the sample should be incubated at 55 °C for 15 min before virus inactivation (9).
Virus inactivation: when collecting specimens, use a virus inactivation preservation tube containing guanidine salt to immediately inactivate the virus; samples taken from the virus preservation solution can be inactivated in a thermostat or water bath at 56 °C for 30 to 45 min (10).
Nucleic acid extraction and loading
Nucleic acids can be manually extracted or automatically extracted. Manual extraction involves many processes with the possibility of cross-contamination, whereas automatic extraction by commercial equipment is highly efficient and safer for operators. It is recommended to use magnetic-bead-based automated extraction. However, “false positives” caused by nucleic acid aerosols should be avoided. Pollution can be minimized by spraying nucleic acid scavengers or 75% alcohol, wiping (not suitable for the magnetic bar of automated extraction instruments), and disinfecting with an ultraviolet lamp.
Nucleic acid amplification
Personnel protection
It is recommended to adopt Biosafety Protection Level 3 when one deals with SARS-CoV-2. If the conditions are limited and the lab staff work in different zones for different tasks, the staff in the amplification zone may wear the PPE required for Biosafety Protection Level 2.
Nucleic acid amplification
Transfer the assembled reactions to the instrument and start the amplification program.
Amplification product treatment
Pack the amplification product tightly in a disposable medical waste bag and transfer it to the amplification product waste disposal area.
Result judgment
Negative
If there is no Ct value or the Ct value is greater than 40, it can be reported as negative.
Positive
If the Ct value is less than 37, it can be reported as positive.
Gray interval
If the Ct value is between 37 and 40, it is recommended to repeat the test. If the Ct value is still less than 40 and the amplification curve has a clear peak, the sample is judged as positive; otherwise, it is negative. If a commercial kit is used, the instructions provided by the manufacturer should be followed.
Laboratory diagnostic criteria
In the “Guidelines for Laboratory Testing of COVID-19” promulgated by the Chinese Health Administrative Agency in March 2020, laboratory-confirmed cases should meet one of the following two conditions:
- Real-time fluorescent PCR results of two SARS-CoV-2 targets (ORF1ab, N) in the same specimen are positive. If a single target is positive, we recommend obtaining a new sample from the same individual and repeating the test. If the result is still positive for a single target, the case is determined to be positive for COVID-19.
- Real-time fluorescent PCR assays of two types of specimen from the same patient both show positive for a single target, or the test results of two samples of the same type both show positive for a single target, the case is determined to be positive for COVID-19. Based on the accumulated data from the testing work in Guangdong Province, Table 2 has been prepared as a reference for the interpretation of test results.

Full table
If both test targets are not specifically amplified, a “negative” can be reported. A negative result may be due to either the viral load being below the testing limit or the lack of a pathogen or nucleic acids in the specimens. It is recommended to combine clinical presentation, imaging examination, and other laboratory tests to prevent misdiagnosis. If clinical signs and other examinations are highly suggestive of SARS-CoV-2 infection, it is recommended to conduct a comprehensive analysis of the entire testing process (such as sample collection, transportation, and processing) and also take into account incubation period, recurrence, or co-infection with other respiratory viruses. If possible, it is recommended to collect samples with better sensitivity, such as deep-cough sputum or alveolar lavage fluid, rather than use nasopharingeal swabs.
Quality control
Standardized clinical nucleic acid testing laboratories have reasonable laboratory zoning, properly trained personnel, well-documented SOP, and comprehensive quality management systems.
Timing and body parts for specimen collection
The viral load in samples may vary depending on the period of infection and the parts of the body from which they were collected. Collecting specimens from multiple parts of the patient’s body may increase the detection rate. As viruses replicate inside of cells, collecting cells containing the virus is one of the most important steps in avoiding “false negatives”.
Specimen collection
Specimen collection workers should be proficient in sample collection. For swab specimens, samples containing more human cells are preferred. An amplification reagent with “internal standard” can be used to check whether the sample is qualified.
Specimen transportation
After the specimen is collected, it should be placed in a special delivery tube containing virus preservation solution or viral transport medium at 4–6 °C and shipped to the laboratory in a timely manner to reduce degradation of the virus RNA during transportation.
Specimen reception and processing
Specimens, especially sputum, should be processed as soon as possible. If the sputum digestive fluid is added to the specimen, a prolonged operation time will result in the degradation of nucleic acids.
Use of quality control materials
CFDA-approved test kits contain positive quality control materials. In addition, confirmed positive nucleic acid from patients may be aliquoted and kept at −80 °C to be used as a positive control for verifying the effectiveness of the reagents. If laboratory conditions permit, positive specimens can be inactivated and used as positive control materials after assessment. Quality control materials should include weakly positive specimens.
Including sufficient quality controls
Molecular assays should include reagent controls, negative controls, and positive controls. It is recommended to include a weak positive control and three negative controls for each batch of SARS-CoV-2 testing. The three negative controls should be randomly placed among clinical samples. If the weak positive control test is positive and all three negative controls are negative, the test is considered to be in control; otherwise, it is considered out of control. The case should be carefully analyzed and re-tested if necessary.
Ensuring the efficacy of the reagents
Quality tests of each batch of reagents with previously identified negative and positive samples can be used to ensure reagent accuracy. Reagents have a “lower testing limit”. If the amount of the virus in a sample is below the detection limit, the virus will not be detected by the reagent.
Serological testing
Specimen types and transferring requirements
Whole blood (including terminal blood), serum, or plasma specimens can be used to test for the specific antibodies of SARS-CoV-2. Existing studies have shown that inactivation of serum at 56 °C for 30 min will affect the test results for the specific antibodies of SARS-CoV-2, especially the IgM antibody test results (11). The positive rate of SARS-CoV-2 antigens in the blood is not yet clear, and urine, nasal, and pharyngeal swabs, sputum, feces, and other specimens can be used for testing if necessary (12). After collecting and packaging the samples, special transport boxes and transport cans should be used to transfer the samples to the laboratory as soon as possible. Relevant departments should appoint particular persons to register, review and distribute the samples, using different boxes for different purposes.
Testing principle
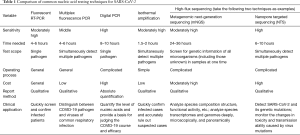
Common methods for serological testing of SARS-CoV-2 include colloidal gold immunochromatography, fluorescence immunochromatography, enzyme-linked immunosorbent assay (ELISA), and the chemiluminescence method (13,14). From a methodological point of view, fluorescence immunochromatography, ELISA, and chemiluminescence are relatively sensitive in testing and can provide semi-quantitative or quantitative test results, which allow for tracking of the dynamic changes in antibody concentrations, but the corresponding detection instruments are required. The results of colloidal gold immunochromatography can be judged by the naked eye. This method is simple and fast, but it has relatively low sensitivity and can only provide qualitative results. A comparison of each testing method is shown in Table 3.

Full table
Interpretation of results
Serological testing of SARS-CoV-2, including tests for antigens and antibodies and tests for the specific antibodies IgM and IgG, has been included in the laboratory testing section of the “COVID-19 Diagnosis and Treatment Protocol” (4). The following can be considered as diagnostic criteria for suspected cases: (I) a positive result for the specific IgM antibody or IgG antibody of SARS-CoV-2, or (II) the specific IgG antibody in serum during the recovery period is at least four times greater than that during the acute period. For confirmed patients, therapeutic effects and the outcome of the disease can be assessed by dynamically monitoring the concentration of SARS-CoV-2 antigens and antibodies as well as serological changes. In addition, testing for SARS-CoV-2-specific antibodies can also be applied to retrospective epidemiological investigations of human infections. It should be noted that the limitations of immunological testing methods regarding antigen-antibody binding, rheumatoid factor (RF), heterophile antibodies, hemolysis, fibrin, and antigen cross-reactions with other types of coronaviruses in the specimen can lead to false positives (15,16). Therefore, serological test results cannot be used independently for the diagnosis of SARS-CoV-2 infection. The patient’s clinical symptoms, exposure history, imaging tests, and other laboratory testing results should also be taken into account to make a comprehensive judgment. See Table 4 for the interpretation of test results for the specific antibodies IgM and IgG.

Full table
Quality control
Kits approved by China’s National Medical Products Administration should be used. Before carrying out testing in the laboratory, relevant performance must be verified according to the performance parameters (such as conformity rate, precision, accuracy) specified by the reagent instructions.
If an external quality assessment agency can provide the corresponding quality control materials, it should participate in an external quality assessment or undergo comparison with other laboratories that carry out the same testing method.
Qualitative testing: For each test batch, it is recommended to use negative and positive quality control materials and to use external quality control. The quality control materials with test result values close to the critical value are positive. If the positive control result is positive and the negative control result is negative, the batch is considered valid. Quantitative testing: Before use each day or after re-calibration, quality control materials provided with the kits or by a third party should be used for quality control, including at least two concentration levels.
Virus isolation and identification
Virus isolation
SARS-CoV-2 can be isolated from human respiratory samples, but this cannot be carried out in general laboratories and therefore is not recommended as a routine diagnostic procedure (17). In a BSL-3 laboratory, specimens such as oral/nasopharyngeal swabs, mouthwashes, nasopharyngeal extracts, tracheal and bronchial secretions, lung biopsy materials, and autopsy tissues are pretreated accordingly. The specimens (200 µL of each) should be tested and inoculated into Vero-E6 or human small airway epithelial cell (HPSAepiC) culture tube and incubated in a 36 °C incubator for 4 h, with gentle shaking every 15 min during the incubation period. The cells should then be washed with 200 µL PBS once, and 1 mL of maintenance solution should be added to continue the culture. When most of the cells show cytopathic effects on the edges (the judgment criteria are shown in Table 5) and irregular morphology, granulating, rounding up, shrinking, and detaching, while the normal control cells are in good shape, the cultured cells should be collected for identification.
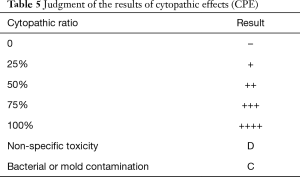
Full table
Virus identification
Fluorescence quantitative PCR, reverse transcription PCR (RT-PCR), indirect immunofluorescence, or electron microscopy are used for observation and identification.
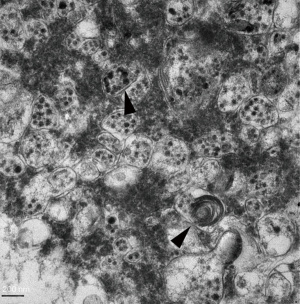
SARS-CoV-2-infected Vero-E6 cells should be incubated for 1 h in a 36 °C incubator and then cultured for 48 h. The infected cells should be scraped with CPE and centrifuged at 3,000 rpm for 10 min at 4 °C. Then, the supernatant should be removed, 1.5 mL of 2.5% glutaraldehyde mixed with 4% paraformaldehyde should be added, and then the solution should be fixed at 4 °C for 2 h. Next, the cells should be rinsed three times with 0.1 M phosphate buffer, fixed with 1% osmium acid, dehydrated by graded ethanol, embedded with the embedding mold Epon-812 pure resin, subjected to conventional ultra-thin sectioning (70 nm), double stained with uranium acetate and lead citrate, and observed through transmission electron microscope (Figures 2 and 3).

Biosafety and waste disposal
SARS-CoV-2 has been included in class B infectious diseases as described in the Law of the People’s Republic of China on the Prevention and Control of Infectious Diseases, but it is treated with the prevention and control measures for class A infectious diseases. The collection, testing, packaging, and transportation of SARS-CoV-2 specimens and the destruction of wastes are subject to the following biosafety requirements.
Biosafety requirements for specimen collection (1)
Specimen collection operators should be trained and qualified in biosafety practices.
Protective equipment of the operators should reach BSL-3 personal protection. If the operators are exposed to a patient’s blood, body fluids, secretions, or excreta, they should change the outer layer of latex gloves at once. If necessary, a waterproof apron or waterproof isolation clothing should be worn.
After collecting the specimens, the outer wall of the container should be disinfected as much as possible before transportation.
Temporary storage of specimens: specimens tested within 24 h can be stored at 4 °C. Specimens that need to be stored for a longer period of time should be stored at −70 °C and below, and repeated freezing and thawing should be avoided.
Biosafety requirements for carrying out experimental activities with specimens (18)
Specimen testing operators should be trained and qualified in biosafety practices.
Specimen sub-packaging
After the specimens are collected, they should be sub-packaged in the BioSafe cabinet of the BSL-2 laboratory. The sub-packaging operators of inactivated specimens are required to adopt BSL-3 personal protection.
Treatment of uncultivated infectious materials
Operations with uncultivated infectious materials should be carried out in a BSL-2 laboratory, and BSL-3 personal protection should be adopted.
Virus culturing
Virus culturing should be carried out in a BSL-3 laboratory. After adding the lysis agent or inactivator to the virus culture, refer to the protection level of the uncultivated infectious material.
Treatment of inactivated materials
Infectious materials or live viruses should be tested in a BSL-3 laboratory after being inactivated by reliable methods.
Biosafety requirements for specimen transportation and packaging (18)
Classification of domestic transport packaging
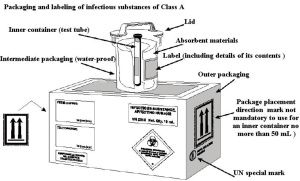
The transport packaging classification of SARS-CoV-2 strains or other potentially infectious biological materials belongs to Class A, and the corresponding UN number is UN2814. The packaging meets the PI602 classification packaging requirements of the ICAO document Doc9284 “Technical Instructions for the Safe Transport of Dangerous Goods by Air”. Transportation of this kind of material shall be carried out in accordance with the “Regulations on the Transport Management of Highly Pathogenic Bacterial (Viral) Strains or Samples That Can Infect Humans” (formerly Ministry of Health Order No. 45). If the same type of highly pathogenic bacterial/viral strain or sample is transported multiple times between an applicant and a receiving organization, it may apply for multiple transports.
Domestic transportation requirements
After a specimen is collected, it should be packaged according to the packaging criteria for infectious substances of class A (see Figure 4); specifically, three-layer packaging is required, which includes: a main container (waterproof, sealed, and leak-proof, such as the test tube containing the specimen), a supplementary container (waterproof, resistant to 95 kPa pressure, leak-proof, such as plastic cans) and an outer packaging (a protective layer outside the supplementary container, with high strength, such as transport boxes or disposable transport packaging). Once packaged with these materials, the specimen should be transported by at least two professionals trained and qualified in biosafety practices in a special vehicle that is equipped with emergency treatment boxes for biohazard overflow. For long-distance transportation, the specimen must be transported on dry ice.
International transportation
SARS-CoV-2 specimens or strains transported internationally should be packaged in a standardized manner. A concerned organization or person should go through the relevant procedures in accordance with the Regulations on the Administration of Health and Quarantine of Special Items for Entry and Exit and meet relevant national and international requirements.
Transportation within an organization
Respiratory and blood samples of SARS-CoV-2 should be sent to the laboratory as soon as possible after collection. In order to avoid accidental leakage or overflow, the specimens can be fixed on the test tube rack to keep the container with the specimens upright. Use sealed special cases, special transportation boxes, tanks, or other containers of level 2 to transport the specimens. It is best to have a gasket at the seal and remove pollution regularly; containers of level 2 can be metal or plastic products, which can withstand autoclaving and chemical disinfectants. Sampling, transportation, and receiving departments need to appoint special persons for registration, review, and transportation.
Specimen and strain management (18,19)
SARS-CoV-2 strains and their samples should be managed by a dedicated person, and a special storeroom or cabinet should be set up to preserve the specimens separately.
The source, type, quantity, and registered number of the strains and samples should be accurately recorded, and effective measures should be taken to ensure the safety of the strains and samples and strictly prevent misuse, malicious use, theft, robbery, loss, leakage, and other events.
Organizations without preserving qualifications should send SARS-CoV-2-positive specimens to those with preserving qualifications or those conducting re-testing within one week after completing the test. If delivery is not required, it should be destroyed within one week.
During the surveillance period of SARS-CoV-2, disease control centers at all levels and BSL-3 laboratories, which are qualified for storage, can be established as temporary storage organizations. If other organizations need to temporarily save and use SARS-CoV-2 samples for scientific research projects or experimental tasks, they may apply to the local administrative department in charge of biosafety.
Disposal of waste (20)
When operating in a BioSafe cabinet, potentially contaminated gloves should be removed in the BioSafe cabinet and thrown in a medical garbage bag in the BioSafe cabinet.
The garbage bag containing medical wastes in biological safety cabinets must be sealed. Medical alcohol (75%) or chlorine-containing disinfectants with effective chlorine 500–1,000 mg/L should be sprayed inside and outside of the seal before transferring it to the yellow medical garbage bags. Before removing the yellow medical garbage bag from the BioSafe cabinet, 75% medical alcohol or chlorine-containing disinfectant with effective chlorine 500–1,000 mg/L should be applied to the seal and the outside of the garbage bag.
If there are splashes or leaks during operation, the pathogenic microorganisms must be killed by disinfection.
All wastes generated during the laboratory operation must be sterilized by pressure steam at 121 °C for 30 min. At the same time, a high-pressure effect indicator should be set. After high-pressure sterilization, the waste can be treated as ordinary waste.
Conclusions and outlook
SARS-CoV-2 is still spreading worldwide. This consensus-based report focuses on the major need for SARS-CoV-2 laboratory testing based on the latest national COVID-19 diagnostic standards and the latest international research progress. It makes use of the rich experience in clinical diagnosis and treatment of the members of the Scientific Research Group of the COVID-19 Prevention and Control Command Office of the Guangdong Provincial Department of Science and Technology to provide cross-application and mutual confirmation, especially regarding the issue of different testing methods leading to different results. Different levels of COVID-19 clinical departments can refer to this report as a reference for diagnosis. With the continuous development of SARS-CoV-2-related diagnostic products and testing methods, this team of laboratory testing experts will continue to work on the applications of new COVID-19 diagnostic technologies and supplement or revise this report in time to meet the needs of COVID-19 laboratory diagnostic testing operators at all levels, so as to improve testing ability and result judgment, detect the source of infection as early as possible, and effectively control the spread of the epidemic.
Acknowledgments
Funding: This work was supported by grants from the National Key Research and Development Program of China (Nos. 2018YFC1200100, 2018YFC1311900, and 2020YFC0842400), the Science Research Project of the Guangdong Province (No. 2019B030316028), and the Guangzhou Medical University High-level University Clinical Research and Cultivation Program (Nos. [2017] 159 and 160).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1928). NZ serves as an unpaid editorial board member of Journal of Thoracic Disease. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kumar S, Maurya VK, Prasad AK, et al. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virusdisease 2020;31:13-21. [Crossref] [PubMed]
- Fung TS, Liu DX. Coronavirus infection, ER stress, apoptosis and innate immunity. Front Microbiol 2014;5:296. [Crossref] [PubMed]
- Mallapaty S. Why does the coronavirus spread so easily between people? Nature 2020;579:183. [Crossref] [PubMed]
- National Health Commission of the People’s Republic of China. COVID-19 Diagnosis and Treatment Protocol (Trial Version 7), [2020-3-3]. Available online: http://www.gov.cn/zhengce/zhengceku/2020-03/04/content_5486705.htm
- van Doremalen NB, Morris T, Holbrook DH, et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med 2020;382:1564-7. [Crossref] [PubMed]
- Chen ZM, Fu JF, Shu Q, et al. Diagnosis and treatment recommendations for pediatric respiratory infection caused by the 2019 novel coronavirus. World J Pediatr 2020;16:240-6. [Crossref] [PubMed]
- National Health Commission of the People’s Republic of China. Guidelines for Laboratory Testing of COVID-19 (Third Edition), [2020-01-28]. Available online: http://www.gov.cn/zhengce/zhengceku/2020-01/29/content_5472893.htm
- Duan XZ, Wang XC, Yu P, et al. The effect of virus inactivation treatment on the weak positive result of SARS-CoV-2 nucleic acid testing. Chinese Journal of Laboratory Medicine 2020. DOI: 10.3760/cma.j.cn114452-20200227-00138. [Crossref]
- Sung H, Yong D, Ki CS, et al. Comparative Evaluation of Three Homogenization Methods for Isolating Middle East Respiratory Syndrome Coronavirus Nucleic Acids From Sputum Samples for Real-Time Reverse Transcription PCR. Ann Lab Med 2016;36:457-62. [Crossref] [PubMed]
- Fischer M, Renevey N, Thür B, et al. Efficacy assessment of nucleic acid decontamination reagents used in molecular diagnostic laboratories. PLoS One 2016;11:e0159274. [Crossref] [PubMed]
- Hu X, An T, Situ B, et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS-CoV-2. J Clin Lab Anal 2020.e23411. [PubMed]
- Diao B, Wen K, Chen J, et al. Diagnosis of Acute Respiratory Syndrome Coronavirus 2 infection by detection of nucleocapsid protein. medRxiv 2020. DOI: 10.1101/2020.03.07.20032524. [Crossref]
- Li Q, Liu DB, Qiao ZR, et al. The value of SARS-CoV-2 IgM / IgG antibody testing in the diagnosis of COVID-19. International Journal of Laboratory Medicine. Available online: http://kns.cnki.net/kcms/detail/50.1176.r.20200304.1041.006.html
- Ning YT, Hou X, Lu MY, et al. Discussion on testing technology application of SARS-CoV-2 specific antibody in serum. Medical Journal of Peking Union Medical College Hospital 2020. DOI: 10.3969/j.issn.1674-9081.20200050. [Crossref]
- Zou MY, Wu GQ. The effect of antigen cross-reaction on testing of SARS-CoV-2 specific antibodies in serum. Chinese Journal of Clinical Laboratory Science 2020;3:161-3.
- Zhang R, Li JM. Talking about false positive testing result of SARS-CoV-2 specific antibodies (IgM/IgG). National Center for Clinical Laboratories, 2020.
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- National Health Commission of the People’s Republic of China. SARS-CoV-2 Laboratory Biosafety Guidelines (Second Edition), [2020-02-13]. Available online: http://www.nhc.gov.cn/qjjys/s7948/202001/0909555408d842a58828611dde2e6a26.shtml
- National Health Commission of the People’s Republic of China. Guidelines for Laboratory Testing of COVID-19 (SIxth Edition) [2020-03-15]. Available online: http://www.gov.cn/zhengce/zhengceku/bmwj/index.htm
- World Health Organization. Laboratory biosafety manual (Third edition). 2004.