
Acute Stanford type B aortic dissection—who benefits from genetic testing?
Introduction
Acute Stanford type B aortic dissections result from a tear in the intimal arterial layer, which allows blood to propagate within the vascular wall. This creates a flap, which divides the aorta into a true and a false lumen. The disease has an incidence of approximately 3–5/100,000 cases per year (1) and—without instant antihypertensive therapy or endovascular intervention—an early mortality rate >50% (2).
The hereditary thoracic aortic aneurysm and dissection (TAAD) syndrome is associated with pathogenic variants in several genes, i.a. fibrillin-1 (FBN1), causing Marfan syndrome, transforming growth factor-β receptor 2 (TGFBR2), causing Loeys-Dietz syndrome and α-actin 2 (ACTA 2), causing isolated thoracic aneurysms and Stanford type B aortic dissections. Approximately 20% of patients with TAAD but absent other organ manifestations (familial non-syndromic TAAD) carry causal single gene mutations with autosomal dominant inheritance (3). A recent review identified 11 genes with definitive TAAD association and 19 further genes with limited or uncertain association (4).
Patients with Marfan syndrome represented 4% of cases in the large international registry of aortic dissection (IRAD trial) (5). However, in dissection patients younger than 40 years of age, the prevalence of Marfan syndrome or related genetic disorders is thought to be much higher. This group of patients, as well as those with a familial inheritance of thoracic aortic disease, should be offered genetic counseling and testing. The finding of pathogenic genetic variants associated with familial aortic disease may have immediate clinical relevance to patients with type B dissections and their first-degree relatives, as reflected in recent recommendations by the European Society for Vascular Surgery concerning counseling, diagnosis and treatment (6). The current study investigated patients with Stanford type B aortic dissection by next generation sequencing after selecting patients on the basis of young age and/or a positive family history of aortic disease.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2421).
Methods
Study design
In this single center cohort study acute Stanford type B aortic dissection were consecutively enrolled between 2013–2018. All patients were of non-Finnish Caucasian ethnicity. The diagnosis was confirmed in all patients by computed tomography angiography (CT-A) using standard radiology protocols. Of all patients, baseline characteristics, risk factors, clinical parameters and treatment data were documented.
Patients were selected for genetic testing if at least one of the following criteria was fulfilled: age of disease onset ≤45 years and/or presence of a first-degree relative with aortic aneurysm or dissection.
Exclusion criteria
Patients with traumatic aortic dissection, known genetic connective tissue disease or history for drug abuse prior to the aortic dissection event were excluded from the study.
Genetic analysis
Exome sequencing was performed at the German Research Center for Environmental Health, Helmholtz Zentrum München, on a Genome Analyzer IIx system (Illumina) after in-solution enrichment of exonic sequences (SureSelect Human All Exon 38 Mb kit, Agilent). Read alignment to the human genome assembly hg19 was performed with the Burrows-Wheeler Aligner (BWA) software, version 0.5.8. Single-nucleotide variants (SNV) were detected with SAMtools (v 0.1.7), as described before (7).
Whole exome sequencing (WES) findings were classified according to the study by Renard et al. as variants in 30 candidate genes with definitive (n=11) or strong (n=19) risk association (ACTA2, COL3A1, FBN1, MYH11, SMAD3, TGFB2, TGFBR1, TGFBR2, MYLK, LOX, PRKG1) and those with moderate or limited association (EFEMP2, ELN, FBN2, FLNA, NOTCH1, SLC2A10, SMAD4, SKI, CBS, COL4A5, PKD1, PKD1, BGN, FOXE3, HCN4, MAT2A, MFAP5, SMAD2, TGFB3) (4). Datasets were also screened for copy number variations (CNV) affecting each of these candidate genes using the FishingCNV algorithm (8). Variant interpretation was performed in accordance with ACGM guidelines and the revised Ghent nosology for the classification of FBN1 variants (9,10).
All patients gave their written consent for genetic testing and study participation. The study was approved by the Medical Ethics Committee of the University Heidelberg, Germany (reference number S-190/2004, amendment S-551/2018). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Genetic counseling and validation were offered to the affected individuals according to the regulations of the German Genetic Diagnostics Act.
Statistical analysis
Fisher’s exact test was performed for the analysis of categorical variables and Students’ t-test for the analysis of continuous variables. The SPSS 19.0 statistics software package was used for statistical analysis.
Results
Out of 105 aortic dissection patients, nine patients (8.6%) were either ≤45 years at the initial diagnosis (n=8) and/or had a first-degree relative with aortic disease (n=2). Blood samples of these nine patients were analyzed for predisposing germline mutations. Compared to the remaining patients of the cohort there was no difference in sex or other reported risk factors such as arterial hypertension and smoking history (Table 1).

Full table
Variant findings in one of the 30 selected candidate genes were prioritized if their coverage (depth) was ≥30 reads and they were found in less than 408 of 20,455 analyzed in-house samples.
Samples were screened for pathogenic variants in genes definitively associated with aortic disease (Table 2, category A). Three patients (patients 2029, 2032, 2008) carried pathogenic variants in FBN1. Two patients showed protein-truncating FBN1-variants c.6366_6367insA, p.(Asp2123GlufsTer5) (patient 2029) and c.4786C>T, p.(Arg1596Ter) (patient 2032). Patient 2008 was heterozygous for a missense substitution c.6661T>C, p.(Cys2221Arg). All three identified FBN1 variants were rare and not found before among 20,455 samples analyzed at the German Research Center for Environmental Health, Helmholtz Zentrum München, Germany. Variant p.(Asp2123GlufsTer5) (patient 2029) was novel, the other two variants were previously detected in patients with Marfan syndrome and classified as “pathogenic/likely pathogenic” by the National Center of Biotechnology Information (NCBI) database of human genetic variation ClinVar.
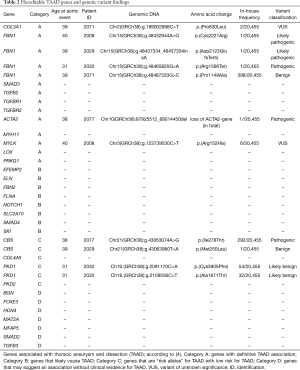
Full table
Patient 2029 died due to aortic dissection complications. Six years after successful type B aortic dissection thoracic endovascular aortic repair (TEVAR) treatment patient 2032 survived type A aortic dissection. Causative for recent ophthalmological syndromes was not an ectopia lentis but a pseudotumor cerebri which is under diagnostic assessment. The patient and family members are not interested in further genetic counselling.
CNV analysis detected a 1,077 kb large deletion on chromosome 10 in patient 2077. This microdeletion encompassed 11 protein-coding genes [(PTEN, FAS partial, NLS, LIPJ, LIPF, LIPK, LIPN, LIPM, ANKRD22, STAMBPL1, ACTA2). One of the genes in the deletion, ACTA2 (actin alpha 2, smooth muscle) was classified as “definitive TAAD risk associated”. This large CNV was therefore classified as likely pathogenic. Recently performed 4-year follow-up CT-A after TEVAR treatment showed no signs of aortic diameter progression without further concomitant diseases or syndromes. Family members haven’t yet presented with aortic syndromes.
In patient 2008 an additional variant in the MYLK (myosin light chain kinase) gene was found, which was classified as variant of unknown significance. Moreover, variants in CBS and PKD1 were found, both genes with limited evidence for association with TAAD (see Table 2), as well as a variant of unknown significance in COL3A1. The patient had a positive family history for aortic syndromes and showed phenotypical system findings of connective tissue disintegration (Ghent Score 8). Follow-up examinations for the uncomplicated chronic type B aortic dissection und er medical therapy revealed no signs of aortic diameter expansion but aortic valve reconstruction was necessary due to relevant insufficiency. No family members are present for further genetic counseling.
Discussion
This single center study explored selection of patients with sporadic type B Stanford aortic dissection for genetic testing, including WES analysis and CNV analysis. Nine out of 105 patients were selected, based on age of disease onset ≤45 years and/ or familial inheritance of thoracic aortic aneurysm and/or dissection (TAAD). Other non-genetic causes for aortic type B dissection in young patients were excluded such as history for blunt traumatic aortic injury or drug abuse. Four of the 9 patients carried pathogenic mutations in TAAD-associated genes. At the time of aortic dissection all four patients with pathogenic genetic findings were aged ≤40 years which is the recommended age threshold for genetic testing in patients with type B aortic dissection (6). In contrast to previous TAAD publications including both aortic aneurysms and dissections, this study investigated exclusively Stanford type B aortic dissections as these patients were referred to the department of vascular and endovascular surgery.
The disease-causing effect of the three pathogenic variants identified in FBN1 seems highly convincing. The DNA-sequence variant rs113871094, identified in patient 2032, results into truncation of the FBN1 protein due to the change of arginine into a stop codon at amino acid position 1,596 of the polypeptide (Arg1596Ter). This variant is not reported in the gnomAD database, a collection of genomes from 141,456 unrelated human individuals from different populations, investigated as part of various disease-specific and population-genetic studies. It was also absent from the 21,455 genomes studied at our center. However, the Arg1596Ter variant in the FBN1 gene has been reported multiple times in association with Marfan syndrome (11-14).
DNA-variant rs113543334, detected in patient 2008 leading to a Cys2221Arg substitution in the FBN1 polypeptide, was also absent from the gnomAD database and collection of genomes studied in our sequencing center. However, the Cys2221Arg missense variant has been reported in the literature in at least three individuals with suspected Marfan syndrome, segregating with disease in two affected family members (15,16).
The rare FBN1 variant that we identified in patient 2029 was a novel finding neither in population-genetic studies, nor in patients with Marfan syndrome or Marfan-like phenotypes. The protein-truncating effect of the variant Asp2123Glu fsTer5, causing premature chain termination due to a frame-shift at amino acid position 2123 of the FBN1 polypeptide, however, makes it highly probable that the variant is pathogenic (17).
The finding of a large genomic deletion in chromosome 10 in patient 2077 is another novel finding of this study. The deletion covered coding sequences of 11 protein coding genes, including ACTA2, the gene encoding alpha-2 smooth muscle specific actin. Mutations in the ACTA2 gene account for approximately 15% of all cases of familial thoracic aneurysms and dissections (TAAD) due to multisystemic vascular smooth muscle cell dysfunction (15-17). Moreover, ACTA2 was classified (together with ACTA1, ACTC1 and ACTG2) as belonging to an ohnolog gene family, implying dose-sensitivity, which makes it likely that loss of the ACTA2 gene has a deleterious effect (http://ohnologs.curie.fr, as at April 2020) (18).
According to the aortic disease guidelines 2014 from the European Society of Cardiology (ESC), TAAD results from either a gene defect affecting the connective tissue, decreased TGF-β signaling or altered vascular smooth muscle cell function. Once a hereditary form of TAAD is suspected, genetic counseling and testing should be offered to the affected patient and family members (class of recommendation I, level of C) (19). TAAD inheritance is mainly autosomal dominant impairing vascular wall integrity by affecting genes (I) for extracellular matrix composition, (II) of the TGF-β signaling pathway and 3) for smooth muscle cell contractility resulting in “phenotypic switching” (20-22).
Next-generation sequencing studies are essential for further categorization of TAAD gene mutations and detection of novel genetic risk variants. It is estimated that specific gene alterations (category B genes) causing enlargement of the thoracic aorta do not inevitably result in aortic dissection (4).
In large series of Stanford type B aortic dissections, a 4% prevalence of Marfan syndrome was estimated (5). The finding of three pathogenic FBN1 variants in this patient cohort suggest that the proposed selection of nine patients for next generation sequencing analysis was an effective strategy to enrich positive genetic testing for Marfan syndrome. Compared to an estimated prevalence of 4% in the unselected type B dissection population, the proposed selection of patients led to a detection of 33% Marfan syndrome patients, which equals a 6–8-fold enrichment compared to an unselected sample. According to Albornoz et al. 50% of aortic dissection patients younger than 40 years of age have Marfan syndrome or a related genetic disorder (23). Supporting previous observations, in this study all affected individuals with pathogenic FBN1 variants were ≤40 years at the time of aortic dissection onset. In accordance to the ESVS guidelines we support genetic testing and counseling in these patients and in individuals with positive history for familial thoracic aortic disease. Reproductive health counseling by genetic specialists should be offered to patients with genetic connective tissue disorder (6). The revised Ghent nosology recommends diagnostic assessment of Marfan syndrome based on family history, aortic root dilation measurement, ectopia lentis and phenotypical systemic features (10). Interdisciplinary medical care should be offered to affected individuals.
Moreover, the detection of three (3% from n=105) patients with Marfan syndrome in the current study sample suggests that patient selection in accordance to the ESVS clinical practice guidelines allows adequate detection of patients with Marfan syndrome in aortic dissection patients. A larger study population with similar selection criteria or control WES studies are needed to precise these results. The “unselected for WES” cohort of this study was not tested for genetical mutations and variations for comparison which is a limitation of the study. Genetic analysis was performed in patients with type B aortic dissection. The existing literature reports different cellular mechanisms for thoracic and abdominal aortic aneurysms and dissection (24). As patients with Stanford type A aortic dissection are referred to the department of cardiac surgery no conclusion can be drawn whether genetic findings are similar to type B aortic dissections. Differentiating between type A, type B aortic dissections and thoracic aneurysms in future genetical studies might be necessary to distinct disease etiologies.
The genotype of TAAD patients presumably determines disease prognosis. Causative genetic variants (“genotype positive TAADs”) showed significant lower aortic event free survival rates compared to patients without any TAAD candidate variants (25).
The genetic findings have therefore direct influence on family members concerning medical counseling, prevention, treatment, surveillance and follow up examinations after aortic dissection. After genetic counseling we recommend medical care for affected patients at interdisciplinary competence centers.
Conclusions
In accordance to the ESVS clinical practice guidelines and the aortic disease guidelines 2014 from the ESC genetic counseling should be offered for patients ≤40 years with sporadic aortic Stanford type B dissection and individuals with suspected familial inheritance for TAAD. This study encourages vascular clinicians to apply molecular-genetic analyses in pre-selected TAAD patients to improve precision medicine.
Acknowledgments
This work was supported by the German Society of Vascular Surgery offering financial support for this study.
Funding: This work was supported by a research grant from the German Society of Vascular Surgery (DGG Forschungsstipendium 2018).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2421
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2421
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2421). PE reports grants from German Society of Vascular Surgery, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of the University Heidelberg, Germany (reference number S-190/2004, amendment S-551/2018). All patients gave their written consent for genetic testing and study participation.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Nienaber CA, Powell JT. Management of acute aortic syndromes. Eur Heart J 2012;33:26-35b. [Crossref] [PubMed]
- Carpenter SW, Kodolitsch YV, Debus ES, et al. Acute aortic syndromes: definition, prognosis and treatment options. J Cardiovasc Surg (Torino) 2014;55:133-44. [PubMed]
- Pinard A, Jones GT, Milewicz DM. Genetics of thoracic and abdominal aortic diseases. Circ Res 2019;124:588-606. [Crossref] [PubMed]
- Renard M, Francis C, Ghosh R, et al. Clinical validity of genes for heritable thoracic aortic aneuryms and dissection. J Am Coll Cardiol 2018;72:605-15. [Crossref] [PubMed]
- de Beaufort HWL, Trimarchi S, Korach A, et al. Aortic dissection in patients with Marfan syndrome based on the IRAD data. Ann Cardiothorac Surg 2017;6:633-41. [Crossref] [PubMed]
- Riambau V, Böckler D, Brunkwall J, et al. Editor's Choice - Management of Descending Thoracic Aorta Diseases: Clinical Practice Guidelines of the European Society for Vascular Surgery (ESVS). Eur J Vasc Endovasc Surg 2017;53:4-52. [Crossref] [PubMed]
- Grond-Ginsbach C, Brandt T, Kloss M, et al. Next generation sequencing analysis of patients with familial cervical artery dissection. Eur Stroke J 2017;2:137-43. [Crossref] [PubMed]
- Shi Y, Majewski J. FishingCNV: a graphical software package for detecting rare copy number variations in exome-sequencing data. Bioinformatics 2013;29:1461-2. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Loeys BL, Dietz HC, Braverman AC, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet 2010;47:476-85. [Crossref] [PubMed]
- Loeys B, Nuytinck L, Delvaux I, et al. Genotype and phenotype analysis of 171 patients referred for molecular study of the fibrillin-1 gene FBN1 because of suspected Marfan syndrome. Arch Intern Med 2001;161:2447-54. [Crossref] [PubMed]
- De Backer J, Loeys B, Leroy B, et al. Utility of molecular analyses in the exploration of extreme intrafamilial variability in the Marfan syndrome. Clin Genet 2007;72:188-98. [Crossref] [PubMed]
- Magyar I, Colman D, Arnold E, et al. Quantitative sequence analysis of FBN1 premature termination codons provides evidence for incomplete NMD in leukocytes. Hum Mutat 2009;30:1355-64. [Crossref] [PubMed]
- Ogawa N, Imai Y, Takahashi Y, et al. Evaluating Japanese patients with the Marfan syndrome using high-throughput microarray-based mutational analysis of fibrillin-1 gene. Am J Cardiol 2011;108:1801-7. [Crossref] [PubMed]
- Schrijver I, Liu W, Brenn T, et al. Cysteine substitutions in epidermal growth factor-like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am J Hum Genet 1999;65:1007-20. [Crossref] [PubMed]
- Attanasio M, Lapini I, Evangelisti L, et al. FBN1 mutation screening of patients with Marfan syndrome and related disorders: detection of 46 novel FBN1 mutations. Clin Genet 2008;74:39-46. [Crossref] [PubMed]
- Faivre L, Collod-Beroud G, Loeys BL, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet 2007;81:454-66. [Crossref] [PubMed]
- Available online: (as at April 2020, Ohnolog family ACTA2 gene).http://ohnologs.curie.fr/cgi-bin/SearchResults.cgi?org_name_search=hsapiens&gene_id=ACTA2&submit_search=
- Erbel R, Aboyans V, Boilea C, et al. ESC guidelines on the diagnosis and treatment of aortic diseases. Eur Heart J 2014;35:2873-926. [PubMed]
- Lin CJ, Lin CY, Stitziel NO. Genetics of the extracellular matrix in aortic aneurysmal diseases. Matrix Biol 2018;71-72:128-43. [Crossref] [PubMed]
- Michel JB, Jondeau G, Milewicz DM. From genetics to response to injury: vascular smooth muscle cells in aneurysms and dissections of the ascending aorta. Cardiovasc Res 2018;114:578-89. [Crossref] [PubMed]
- Ostberg NP, Zafar MA, Ziganshin BA, et al. The Genetics of Thoracic Aortic Aneurysms and Dissection: A Clinical Perspective. Biomolecules 2020;10:182. [Crossref] [PubMed]
- Albornoz G, Coady MA, Roberts M, et al. Familial thoracic aortic aneurysms and dissections - incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg 2006;82:1400. [Crossref] [PubMed]
- Quintana RA, Taylor WR. Cellular mechanisms of aortic aneurysm formation. Circ Res 2019;124:607-18. [Crossref] [PubMed]
- Poninska JK, Bilinska ZT, Franaszczyk M, et al. Next-generation sequencing for diagnosis of thoracic aortic aneurysms and dissections: diagnostic yield, novel mutations and genotype phenotype correlations. J Transl Med 2016;14:115. [Crossref] [PubMed]


