Qualitative research of alternatively splice variants of fibronectin in different development stage of mice heart
Introduction
Embryonic heart development is a very complicated process, which is not only controlled by regulation of temporal and spatial gene expression, and is also involved with cell migration, differentiation, proliferation, and cell-cell interactions. The occurrence of congenital heart disease (CHD) is an abnormal genetic expression process regulated at a variety of levels (1-3). Cardiac neural crest (CNC) plays a requisite role during cardiovascular development. Migration as well as expansion and maintenance of the CNC cells are important for the normal development of the embryonic heart. Fibronectin (FN) protein is present along neural crest migration paths; FN and integrin α5 modulate CNC proliferation and survival, and are required for the presence of CNC cells (4). FN plays vital roles in cell adhesion, differentiation, proliferation, migration and signal transduction. It is involved in processes of embryonic development and is highly conserved in evolution (5,6).
As an important extracellular matrix glycoprotein existing only in vertebrates, FN contains multiple copies of three repeating regions: type I, II, and III. FN diversity is obtained by alternative splicing of EIIIA, EIIIB and IIICS. The EIIIA and EIIIB of FN show a very high degree of homology among vertebrates, and are cassette-type exons which can be independently spliced-in or out from the pre-mRNA, whereas that of the IIICS region is species-specific, five variants in humans, three in rodents, and two in chickens (7). The alternative splicing of FN1 gene introduces another level of complexity and raises questions such as whether the inclusion or exclusion of the alternatively spliced domains alters the functions of FN and the biological consequences. Some in vitro studies have claimed various, but often inconsistent effects of inclusion of EIIIA or EIIIB exons on cell attachment, migration, proliferation, cell survival, matrix assembly, or expression of smooth muscle α actin (8-10). However, the individual deletion of EIIIA or EIIIB has not given much insight into the possible functions of these splice variants in vivo. EIIIA-null or EIIIB-null mice are vital and fertile, and embryonic vascular development and retinal angiogenesis after birth proceeded apparently normal in these mice (11,12). EIIIA-null mice grew normally, though showed alleviation of atherosclerosis (13). Interestingly, a possible effect of EIIIB on FN matrix assembly and proliferation was observed in EIIIB-null mouse embryonic fibroblasts (MEFs) in vitro, suggesting a possible role for EIIIB in FN matrix assembly (12).
Special structure, complicated splice variants, and multiple domains of FN lead to its various biological function. FN1 gene participates in embryonic heart development, through which exons EIIIA and EIIIB may play a vital role. The expression level and ratio of FN1 splice variants may change accordingly in embryonic development and some pathological processes. We aimed to probe existence and variation of FN1 splice variants in heart development. Because FN1 gene is highly conservative during evolution, we chose C57 mice in the present study.
Subjects and methods
Subjects
Adult C57 mice were bought and raised by the animal center of Beijing University, and selected with the female and male at ratio of 1:1. After successful mating, the earliest time that vaginal suppository shed from the female mice was counted for embryonic 0 day (E0d), and the earliest time that the neonatal mouse was born for postnatal 0 day (P0d). Embryos at E11.5d, E12.5d, E13.5d, E14.5d, E15.5d, E16.5d, E17.5d, E18.5d, E19.5d, P1d, and adult male mice (3 months) were selected, and 4 mice were prepared at each time point. All the procedure was accorded with animal ethical standards and under supervision of Institutional Animal Care and Use Committee (IACUC) of Beijing University, and acquired international qualification of Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC), animal experiment IACUC number: IMM-TianXL-02.
Total RNA extraction and RT-PCR
Mice were killed and hearts were taken out and dipped into High Potassium Tyrode, cleaned by PBS, then some myocardial tissue of ventricular septum were cut off. Trizol reagent (Beijing Biomed Co. Ltd China) was employed to extract total RNA of myocardial cell according to manufacturers’ instruction. RNA concentration was measured by NanoDrop, and total RNA was preserved in −80 °C condition. M-MLV revertase, Random Hexamers Primer and RNasin (Beijing TransGen Biotech Co. Ltd China) was used to obtain cDNA as instructed. cDNA was carefully preserved in −20 °C condition.
According to FN1 gene sequence (NM_010233.2) from NCBI data bank, we designed three pairs of primers for alternatively spliced FN1 domains (EIIIB, EIIIA and IIICS). Primer sequences were showed in Table 1. Primers were synthesized by Beijing Huada gene Co. Ltd. PCR amplification was performed in a 25 µL reaction volume including 10× PCR buffer L 2.5 µL, 2.5 mM dNTP 0.5 µL, 10 µM inner primer 0.5+0.5 µL (synthesized by Beijing Huada gene Co. Ltd China), Taq DNA Polymerase (5 U/µL) 1 µL, cDNA 5 µL, ddH2O 15 µL. PCR condition was: preheated in 95 °C (3 min), 35 circles of 95 °C (30 s), 55 °C (45 s), and 72 °C (45 s), followed by 72 °C (2 min) and 4 °C (5 min).

Full table
Electrophoretic identification of PCR products
Agarose gel electrophoresis was adopted to identify RT-PCR products, with electrophoresis apparatus parameter: 200 mA, 150 V, 25 min. Electrophoresis fragments of different size determined whether alternatively spliced domains were include in FN1 gene or not. Specific electrophoresis fragments were showed as follows: EIIIA+ 573 bp, EIIIA− 303 bp; EIIIB+ 538 bp, EIIIB− 265 bp; IIICS+ 569 bp, IIICS− 236 bp.
Results
The results are shown in Figures 1,2. Each cDNA sample was pooled from four mice and was amplified with three pairs of primers, respectively. For each time point, PCR were products amplified with primers of EIIIA, EIIIB, and IIICS, respectively. Specific electrophoresis fragments were showed as the followings: EIIIA+ 573 bp, EIIIA− 303 bp; EIIIB+ 538 bp, EIIIB− 265 bp; IIICS+ 569 bp, IIICS− 236 bp. The makers from top to bottom were: 1,500, 1,000, 900, 800, 700, 600, 500, 400, 300, 200 and 100 bp.
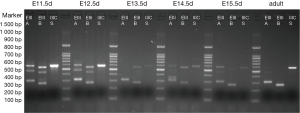
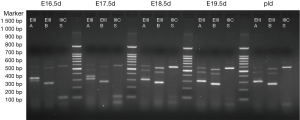
The results revealed that, in embryonic mice, alternatively splice variant of EIIIA+/−, EIIIB+/−, IIICS+ was observed from E11.5d to P1d, whereas in adult mice myocardial cell only alternatively splice variant of EIIIA−, EIIIB−, IIICS+ was expressed (diminished lanes of 573 and 538 bp). IIICS was found in both myocardia of embryonic and adult mice, and EIIIA and EIIIB were only expressed in embryonic myocardial cell of mice. Intriguingly, EIIIA and EIIIB seemed to be highly expressed in early stage of embryonic heart development, implying their crucial role in certain stage of cardiomyogenesis.
Discussion and conclusions
Previously, to identify genes relevant to ventricular septal defects (VSD), we have undertaken suppression subtractive hybridization (SSH) and got possible candidate genes. Then we established cardiomyocytes differentiation model utilizing P19cl6 cell and successfully differentiated them into beating cardiomyocytes with special mark genes in vitro. By detecting genes expression in different development period, thirteen candidate genes were obtained, including FN1 gene. Our study demonstrated that FN1 gene is highly expressed in process of myocardial cell differentiation, possibly participates in embryonic heart development, and plays a vital role in development of CHD including VSD (14).
Embryonic cardiovascular development is a very complicated process, which is involved with cell migration, differentiation, proliferation, and cell-cell interactions. FN is crucial for normal development of embryonic heart by modulating CNC proliferation and survival, and maintenance of CNC cells. FN has been found only in vertebrates, and its appearance in evolution correlates with the emergence of organisms with endothelial cell-lined vasculature (15). Another study by Hanson and colleagues revealed that, in murine heart at embryonic stages, FN was present in the epicardium, with highest levels at E12.5 and present in the myocardium and the endocardium at relatively constant levels at all-time points from E12.5 to P2 (16).
EIIIA/EIIIB double null mice display a reduced number of α-SMA-positive cells immediately surrounding the dorsal aorta, possibly caused by a delay in the recruitment or differentiation of these cells (17). The present study demonstrated that EIIIA and EIIIB domain of FN were expressed only in embryonic hearts of mice. FN1 gene seems to play a significant role by expression of such two highly conserved alternatively spliced domains. Some previous studies unexpectedly revealed that mice with single deletions of EIIIB or EIIIA exons were viable and fertile (11-13). Astrof and colleagues reported that EIIIA/EIIIB double-null embryos displayed multiple embryonic cardiovascular defects, including vascular hemorrhage, failure of remodeling embryonic and yolk sac vasculature, defective placental angiogenesis and heart defects, and showed embryonic lethality with incomplete penetrance by embryonic day 10.5. The absence of EIIIA and EIIIB leaded to diverse cardiovascular defects ranging from severe to mild malformations and some progeny survive. Most of the defective embryos showed various forms of heart defects including inflated pericardial cavity, pericardial edema, degenerating linear heart tube, or dilated aortic sac. E9.5d embryos displaying heart defects were most severely affected. E10.5d embryos showed cushion defects (17), suggesting the critical role for EIIIA and EIIIB in the epithelial to mesenchymal transition in embryonic heart. Because FN deleting EIIIA and EIIIB showed no deterring of integration to cell surface receptor, the embryonic lethality could be due to deletion of EIIIA and EIIIB. Here we showed that EIIIA and EIIIB had differential expression chronologically in embryonic hearts of mice and were especially overexpressed in E11.5d and E12.5d, indicating the dynamic change of expression in specific heart development stage. FN1 gene could participate in embryonic cardiovascular development, through which exons EIIIA and EIIIB play a crucial role.
The absence of FN affects organ assembly but not specification of precursor cells in mice and zebrafish of FN-null mutations. While the molecular mechanisms of these defects are not completely clarified, the absence of FN alters cell polarity leading to defects in polarized cell movements during cell migration in fish and frogs (18-21). Cardiovascular defects in FN-null and EIIIA/EIIIB-null embryos could also be a consequence of defects in endothelial cell polarity, resulting in aberrant endothelial tube formation. The high level of conservation of the EIIIA and EIIIB sequences of FN during evolution and of their tightly regulated patterns of splicing, in contrast with their low homology within same species (only 28% homology between human EIIIA and EIIIB amino acid sequences), strongly suggested a distinct role of these domains. While EIIIA and EIIIB may involve in the same biological process, their functions could be very different. Additionally, a recent study by Franz and colleagues showed that, a relevant re-occurrence of EDA(+) FN and B(+) Tn-C following human heart transplantation could be demonstrated with spatial association to signs of rejection and a significant correlation to tissue inflammation, indicating the role of FN in the cardiac rejection process (22). This is in concert with another study by Booth et al., showing that while EDA(-/-) mice developed acute cardiac rejection in a manner indistinguishable from WT controls, cardiac allografts in EDA(-/-) mice were protected from fibrosis associated with chronic rejection, i.e., EDA cFN promotes the development of fibrosis associated with chronic rejection (23).
These splice variants are expressed at very low levels or not at all in the adult, as we found in adult mice. During embryonic development, EIIIA and EIIIB are expressed around embryonic heart and vessels; but such expression ceases when the process of cardiovascular development is complete. While in adults, splice variants including EIIIA and EIIIB appear only during pathological processes requiring growth or remodeling of the vasculature, for example in the process of tumor development, atherosclerosis, and myocardial infarction these exons become included into FN (13,24-26). Briefly, they show a tight tissue-specific expression, adult animals tend to be devoid of these extra domains (27).
Indeed, the presence of at least one of the extra domains is required for normal cardiovascular development. Furthermore, it appears that EIIIA and/or EIIIB cFN is necessary for the recruitment or differentiation of vessel associated α-SMA cells. FN containing EIIIA and/or EIIIB could be critical in regular endothelial cell polarity and normal endothelial tube formation. Conclusively, FN, and more specifically the EIIIA and EIIIB domains are critical for embryonic cardiovascular development. Further studies with manipulation of FN transcription variants, particularly EIIIA and EIIIB, at both cellular and whole body levels, would provide new insights into the role of FN in cardiac development and the mechanisms behind.
Acknowledgements
Funding: The study was supported by National Natural Science Foundation of China (81370280, 81570332 and 81470544).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Keyte A, Hutson MR. The neural crest in cardiac congenital anomalies. Differentiation 2012;84:25-40. [PubMed]
- Mostafavi-Pour Z, Askari JA, Whittard JD, et al. Identification of a novel heparin-binding site in the alternatively spliced IIICS region of fibronectin: roles of integrins and proteoglycans in cell adhesion to fibronectin splice variants. Matrix Biol 2001;20:63-73. [PubMed]
- Liao YF, Gotwals PJ, Koteliansky VE, et al. The EIIIA segment of fibronectin is a ligand for integrins alpha 9beta 1 and alpha 4beta 1 providing a novel mechanism for regulating cell adhesion by alternative splicing. J Biol Chem 2002;277:14467-74. [PubMed]
- Mittal A, Pulina M, Hou SY, et al. Fibronectin and integrin alpha 5 play essential roles in the development of the cardiac neural crest. Mech Dev 2010;127:472-84. [PubMed]
- Danen EH, Yamada KM. Fibronectin, integrins, and growth control. J Cell Physiol 2001;189:1-13. [PubMed]
- Müller EJ, Williamson L, Kolly C, et al. Outside-in signaling through integrins and cadherins: a central mechanism to control epidermal growth and differentiation? J Invest Dermatol 2008;128:501-16. [PubMed]
- White ES, Baralle FE, Muro AF. New insights into form and function of fibronectin splice variants. J Pathol 2008;216:1-14. [PubMed]
- Chen W, Culp LA. Adhesion mediated by fibronectin’s alternatively spliced EDb (EIIIB) and its neighboring type III repeats. Exp Cell Res 1996;223:9-19. [PubMed]
- Hashimoto-Uoshima M, Yan YZ, Schneider G, et al. The alternatively spliced domains EIIIB and EIIIA of human fibronectin affect cell adhesion and spreading. J Cell Sci 1997;110:2271-80. [PubMed]
- Serini G, Bochaton-Piallat ML, Ropraz P, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol 1998;142:873-81. [PubMed]
- Muro AF, Chauhan AK, Gajovic S, et al. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J Cell Biol 2003;162:149-60. [PubMed]
- Fukuda T, Yoshida N, Kataoka Y, et al. Mice lacking the EDB segment of fibronectin develop normally but exhibit reduced cell growth and fibronectin matrix assembly in vitro. Cancer Res 2002;62:5603-10. [PubMed]
- Tan MH, Sun Z, Opitz SL, et al. Deletion of the alternatively spliced fibronectin EIIIA domain in mice reduces atherosclerosis. Blood 2004;104:11-8. [PubMed]
- Zhang H, Zhou L, Yang R, et al. Identification of differentially expressed genes in human heart with ventricular septal defect using suppression subtractive hybridization. Biochem Biophys Res Commun 2006;342:135-44. [PubMed]
- Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol 2000;150:F89-96. [PubMed]
- Hanson KP, Jung JP, Tran QA, et al. Spatial and temporal analysis of extracellular matrix proteins in the developing murine heart: a blueprint for regeneration. Tissue Eng Part A 2013;19:1132-43. [PubMed]
- Astrof S, Crowley D, Hynes RO. Multiple cardiovascular defects caused by the absence of alternatively spliced segments of fibronectin. Dev Biol 2007;311:11-24. [PubMed]
- George EL, Georges-Labouesse EN, Patel-King RS, et al. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 1993;119:1079-91. [PubMed]
- Marsden M, DeSimone DW. Regulation of cell polarity, radial intercalation and epiboly in Xenopus: novel roles for integrin and fibronectin. Development 2001;128:3635-47. [PubMed]
- Marsden M, DeSimone DW. Integrin-ECM interactions regulate cadherin-dependent cell adhesion and are required for convergent extension in Xenopus. Curr Biol 2003;13:1182-91. [PubMed]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell 2004;6:371-82. [PubMed]
- Franz M, Matusiak-Brückner M, Richter P, et al. De novo expression of fetal ED-A(+) fibronectin and B (+) tenascin-C splicing variants in human cardiac allografts: potential impact for targeted therapy of rejection. J Mol Histol 2014;45:519-32. [PubMed]
- Booth AJ, Wood SC, Cornett AM, et al. Recipient-derived EDA fibronectin promotes cardiac allograft fibrosis. J Pathol 2012;226:609-18. [PubMed]
- Ffrench-Constant C, Van de Water L, Dvorak HF, et al. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol 1989;109:903-14. [PubMed]
- Matter CM, Schuler PK, Alessi P, et al. Molecular imaging of atherosclerotic plaques using a human antibody against the extra-domain B of fibronectin. Circ Res 2004;95:1225-33. [PubMed]
- Shekhonin BV, Guriev SB, Irgashev SB, et al. Immunofluorescent identification of fibronectin and fibrinogen/fibrin in experimental myocardial infarction. J Mol Cell Cardiol 1990;22:533-41. [PubMed]
- Pagani F, Zagato L, Vergani C, et al. Tissue-specific splicing pattern of fibronectin messenger RNA precursor during development and aging in rat. J Cell Biol 1991;113:1223-9. [PubMed]


