Proteomic profiling differences in serum from silicosis and chronic bronchitis patients: a comparative analysis
Introduction
Lungs are essential for respiration, and the mucosal lining is crucial for gaseous exchange. However this lining can be affected by external factors such as microbes, chemical fumes and mineral dusts. Pulmonary diseases including asthma, chronic bronchitis (CB), and chronic obstructive pulmonary disease (COPD) were widespread. CB was characterized by recurrent excessive mucous secretion in the bronchial tree and was accompanied by a coughing and sputum production for at least three months per year for two consecutive years (1,2). The clinical consequences of CB included declining lung function and possible failure, and a deteriorating quality of life (3). The pathology of CB included mucous metaplasia that occurred in response to inflammatory signals, and excessive mucus production by goblet cells (1).
Acute or chronic viral or bacterial infection, and inflammatory cell activation resulted in hypersecretion of mucus that was compounded by decreased ciliary function, distal airway occlusion and ineffective coughing (4). Mucous metaplasia led to airflow obstruction due to luminal occlusion caused by increased mucus hypersecretion, epithelial layer thickening encroaching on the airway lumen, airway surface tension alteration and predisposition to expiratory collapse. Inflammation-associated cells such as T-cells, which are mainly divided into Th1 and Th2 subgroups, produce interleukins IL-1, IL-4, IL-6 and IFN-γ that were believed to be responsible for mucous metaplasia (5). Effector T-cells were a distinct subgroup of CD4+ T-cells such as TH17 cells that possessed potent pro-inflammatory activity (6). A numbers of therapeutic approaches had been developed based on different pathophysiological mechanisms that decreased mucus production and hypersecretion by controlling inflammation from coughing (1). Therapeutic interventions for CB included cessation of smoking if applicable, physical measures such as chest physiotherapy (PT), expectorants and mucolytics, methylxanthine asthma drugs, short- and long-acting β-adrenergic receptor agonists, anticholinergics, glucocorticoids, antioxidants and antibiotics.
Although a severe occupational pulmonary disease, the detailed mechanisms underpinning the pathology of silicosis remained elusive, but it was widely accepted that chronic inflammation was involved in the development of pulmonary dysfunction. Exposure to dust initiated a sequence of events that led to the accumulation and activation of inflammatory cells in respiratory tract (7), proliferation of fibroblasts, and an increase in extracellular matrix components (8). Alveolar macrophages (AMs) played a pivotal role in the pathogenesis of silicosis by releasing various mediators such as destructive proteolytic enzymes and inflammatory growth and differentiation factors (9). Residual endothelial, epithelial and fibroblast cells secrete and express cytokines and other molecules involved in the inflammatory and fibrotic process (10). Dust particles inhalation triggered AMs to release oxygen radicals and eicosanoids, and TNF and IL-6 were also increased in silicosis patients by AMs, where they played a crucial role in inflammatory and fibrosis (11). TNF induced the recruitment of inflammatory lymphocytes, neutrophils and eosinophils, and enhances the expression of chemotactic factors that stimulate fibroblast chemotaxis and growth during development of fibrosis (12). IL-6 was an important inflammatory factor that induced cellular adhesion of monocytes and facilitates infiltration into lung. Il-6 may be involved in the autoimmune process in association with TNF and IL-1, and was implicated in the fibrotic response (13). Release of platelet-derived growth factor (PDGF) and insulin-like growth factors from AMs was reported to induce competence and progression signals during fibrosis (14), and expression of PDGF, TGF and TGF-β are enhanced in the epithelial lining fluid (ELF) of pneumoconiotic patients (8). TGF-β induced the production of PDGF, stimulates fibroblasts (at low concentrations), down-regulates PDGF receptor expression and inhibits fibroblast proliferation (at high concentrations). TGF-β also induced the synthesis of collagen type I and III (15). Investigating differentially expressed proteins involved in cytokine production and signaling may help to explain severe pulmonary lesions associated with silicosis.
The mechanisms underlying the pathophysiology of silicosis and CB were poorly understood at present. In this study we performed proteomic profiling of serum form silicosis and CB patients by using 2-D electrophoresis and MALDI-TOF-MS (matrix assisted laser desorption ionization time of flight mass spectrometry) approaches. Differentially accumulated proteins were identified and analyzed by using gene ontology (GO) classification and various bioinformatics tools. Enzyme linked immunosorbent assay (ELISA) was subsequently used for validation and quantitative analysis of protein and peptide expression data. These results provided new insight into the pathogenesis of silicosis and CB.
Materials and methods
Sample collection
Three groups of whole blood samples were collected from stage I & II silicosis (SI and SII) and CB patients at the 8th People’s Hospital of Wuxi (Wuxi, China). Each group contained 15 individuals and their ages were from 55–64 years old (no significantly difference between three groups). For the patients of SI, SII and CB, each group was diagnosed exclusively and people who suffered from no other diseases were included. Permission for these studies was obtained from the medical ethics committee and all the subjects provided written informed consent using protocols that complied with the Declaration of Helsinki Principles. All the blood samples were stored at 4 °C for 24 h, centrifuged at 4,000 rpm for 10 min (5418R, Eppendorf, German), and serum was stored at −80 °C for a few days until used.
Protein extraction and removal of highly abundant proteins
In order to increase the reliability, five samples within groups were pooled into a new sample, consequently each group was performed in triplicate. Total proteins were extracted by using trichloroacetic acid, and highly abundant proteins were removed by using Agilent human 14 multiple affinity removal system columns (Agilent, USA) following the manufacturer’s instructions. This kit was specifically designed to remove albumin, IgG, antitrypsin, IgA, transferrin, haptoglobin, fibrinogen, alpha2-macroglobulin, alpha1-acid glycoprotein, IgM, apolipoprotein AI, apolipoprotein AII, complement C3, and transthyretin. Selective immunodepletion provided a replenished pool of low-abundance proteins for proteomic analysis with improved resolution and dynamic range for 2D-gel electrophoresis and mass spectrometry. Serum samples were subsequently dissolved in lysate solution (9 M urea, 4% CHAPS, 65 mM DTT and enzyme inhibitor cocktail) and quantified using Coomassie brilliant blue.
2D-gel electrophoresis
Samples (200 ng) in hydration solution (8 M urea, 2% CHAPS, 65 mM DTT, 0.50% IPG buffer) was loaded onto Immobiline Drystrip gels (18 cm, pH 3–10, GE Healthcare, USA) on an IPG-phor system (Amersham Bioscience, USA) for first dimensional electrophoresis. Isoelectric focusing (IEF) was conducted at 30 V for 6 h, 60 V for 6 h, 500 V for 1 h, 1,000 V for 1 hand 8,000 V for 20 h (64,000 Vhr in total). IEF gels were transferred to balanced solution (50 mM Tris HCl, 6 M urea, 2% SDS), agitated for 15 min, and sealed with low-melting point agarose. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed at 4 W for 45 min followed by 15 W until the bromophenol blue reached the bottom of the gel on an Ettan DALT SIX vertical system (Amersham Bioscience, Sweden). SI, SII and CB samples were analyzed in triplicate. After visualization by sliver nitrate staining, gels were scanned by using an Artix Scan 1010 plus (Microtek, Taiwan), and analyzed by Image Master 2D platinum software (GE Healthcare, USA). Protein spots exhibiting at least a two-fold difference were quantified, background-subtracted and filtered, according to two folds change criteria (Shanghai Yeslab Biotechnology, China).
Matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF-MS)
Selected protein spots were excised, transferred to a 96-well plate, sonicated for 5 min. de-stained with acetonitrile, and dried in vacuum for 5 min. Samples were then heated in a water bath at 56 °C with 10 mM DTT and 50 mM NH4HCO3 for 1 h. After addition of 55 mM iodoacetamide and 25 mM NH4HCO3 at room temperature, samples were incubated in the dark for 45 min, washed in 25 mM NH4HCO3, 50% acetonitrile and finally 100% acetonitrile. Trypsin (Promega, USA) was diluted with 25 mM NH4HCO3, and samples incubated on ice for 30 min, then were covered with NH4HCO3 and incubated at 37 °C for 24 h. Proteolysis was terminated with 0.1% trifluoroacetic acid (TFA).
Tryptic peptides were identified by using an Autoflex II MALDI-TOF/MS (BRUKER, Germany) in positive mode, and peptide mass fingerprint (PMF) maps were analyzed by Flexanalysis 3.0 with the SNAP algorithm and a signal-to-noise ratio of 1.5 (Bruker Daltonik, Germany). The Mascot search engine from Matrixscience (www.matrixscience.com) used PMF data were searched against the NCBI redundant database with a trypsin cleavage parameter setting, a peptide mass tolerance of ±0.2 Da, and a maximum of one missed cleavages.
Enzyme linked immunosorbent assay (ELISA)
Expression of Granzyme A, apolipoprotein and Cathepsin-G in the serum of silicosis and CB patients was measured by using ELISA. SI, SII, and CB serum samples were placed into ELISA plates (Yeslab Biotechnology, China), closed with plate closure membranes, and incubated for 30 min at 37 °C. After five washes with washing buffer, 50 μL of HRP-conjugated reagents were added to each well and plates incubated for 30 min at 37 °C, washed five more times, and 50 μL of Chromogen solutions A and B were added, mixed, and incubated for 15 min at 37 °C in the dark. Stop solution was added to, and plates were assayed in a microplate reader (Multiskan FC, Thermo Scientific, Finland) at 450 nm. Standard density curves were plotted, according to the manufactures protocol and the sample density was calculated using the curve equation.
Data analysis
Differentially accumulated proteins identified by using 2D-gel electrophoresis were log-transformed and hierarchically clustered by centroid linkage by using Cluster V3.0 and visualized using clustergrams in Java TreeView (V1.1.6r2) (16). Sequences of identified proteins were searched against the database of non-redundant protein sequence with entries from NCBI RefSeq by Blast2GO (V2.7.2) for gene-ontology annotation (17). Differentially expressed proteins in SI, SII and CB samples were analyzed by method of T-test (SAS 9, USA).
Results
Proteomic profiling of silicosis and chronic bronchitis
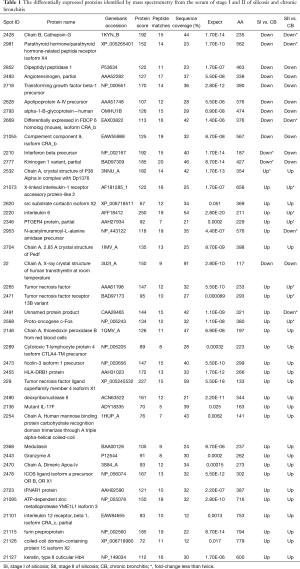
The proteomes of serum obtained from stage I and II silicosis and CB patients were analyzed by 2D-gel electrophoresis in a non-linear pH range of 3–10. In total, 369 protein spots were detected, among which 41 were successfully identified by mass spectrometry (Figure 1; Table 1).
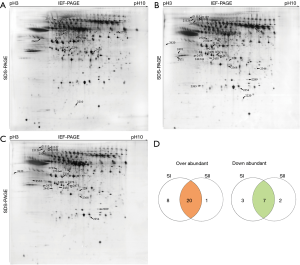
Full table
In total, 28 and 21 proteins were up-regulated in SI and SII, respectively, while 10 and 9 were down-regulated, compared with CB (Figure 1A,B), among which, 20 were up-regulated in both SI and SII, and 7 were down-regulated in both silicosis groups (Figure 1A-D). Details of the identified proteins including protein score, sequence coverage and molecular weight are shown in Table 1.
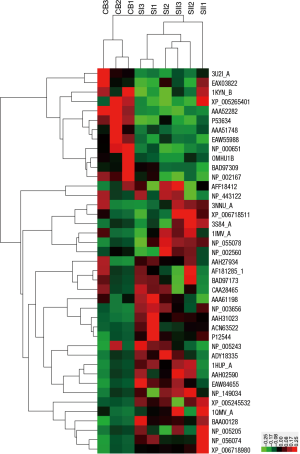
Differentially expressed proteins were hierarchically clustered based on Pearson correlation and visualized by using Treeview software (16). Serum samples mainly clustered into two groups (silicosis and CB) in the top tree of the dendrogram, which implied that the results within groups were highly consistent (Figure 2). From the left tree structure, differentially expressed proteins mainly clustered into up- and down-regulated proteins (Figure 2).
Gene ontology enrichment of differentially expressed proteins
Based on the protein cluster analysis, identified proteins divided into up- and down-regulated categories (Figure 2), and all proteins were annotated using Blast2GO (17). GO of differently expressed proteins was analyzed according to the biological process, molecular function and cellular component categories. At the biological process category, for the up-regulated proteins in SI and SII compared with CB, the top five enrichment annotation-terms were single-organism process, cellular process, biological regulation, response to stimulus and metabolic process (Figure 3A). The cellular component class for these proteins were cell, membrane, membrane-bound organelle, extracellular region and protein complexes, which accounted for 68% of this class annotation (Figure 3B). The top four enriched terms covered the functions of binding, catalytic activity, receptor activity and molecular transducer activity, which accounted for 85% of the molecular function classification (Figure 3C).
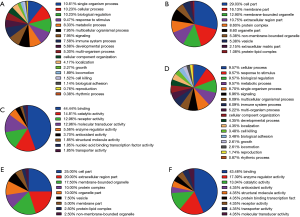
For down-regulated proteins in silicosis patients, the top six enriched terms in biological process class were cellular process, response to stimulus, biological regulation, metabolic process, single-organism process and signaling, which accounted for over 50% of this class (Figure 3D). Cellular component class mainly consisted of cell, extracellular region, membrane-bound organelle, protein complex and organelle, and these enrichment terms were accounted for 82% in this category (Figure 3E). In the molecular function category, the top three terms were binding, enzyme regulator activity, and catalytic activity, which accounted for 74% (Figure 3F).
Validation of selected candidates by enzyme linked immunosorbent assay (ELISA)
Of the identified proteins, three (Granzyme A, apolipoprotein and Cathepsin-G) were selected for validation by ELISA, and the expression levels of all the three were consistent with the 2D-gel electrophoresis results (Figure 4); Expression of apolipoprotein and Cathepsin-G were down-regulated in SI and SII by using both approaches (Figure 4A,C), while Granzyme A was up regulated in both SI and SII according to both 2D-PAGE and ELISA (Figure 4B).

Discussion
The detailed molecular mechanisms unpinning the pathology and physiology of silicosis and CB remain poorly understood. In this study we performed proteomic profiling on serum samples from silicosis and CB patients, and found that cytokines, glycoproteins, metabolism and immunity-related proteins were dramatically up- or down-regulated (Figure 1; Table 1).
Cytokines
The TGF beta family were multifunctional peptides that regulate proliferation, differentiation, adhesion, migration and other functions in various cell types, and the TGF-beta1 precursor was up-regulated in CB compared with silicosis samples (Table 1; Figure 1). The TGF-beta1 signaling pathway may play an important role in the development of fibrosis in autoimmune liver diseases (18). TGFB receptors were presented in many different cell types, and TGFB positively and negatively regulates numerous other growth factors and itself frequently was up-regulated in tumor cells, and mutations in TGFB resulted in Camurati-Engelmann disease. Using animal models, smoking was shown to induce over-expression of TGF-beta1 in bronchial epithelia, and this molecule may be involved in the pathogenesis of smoking-induced CB and emphysema (19). Compared with silicosis, upregulation of TGF-beta1 precursor in CB suggested that TGF-beta1 signaling differed in these two diseases.
Interferon beta (IFN-β) was a highly active cytokine produced in large quantities by fibroblasts that have antiviral activity and were involved in the innate immune response. Type I IFN (IFN-α and IFN-β) signals via a heterodimeric receptor consisting of IFNAR1 and IFNAR2, and recent research showed that type I IFN mediates pulmonary arterial hypertension via IFNAR1 (20). Type I IFN played an important role in the development of crystalline silica-induced lung inflammation in mice, indicating that viruses and inorganic particles trigger similar signaling pathways (21). In response to silica inhalation, IFN-β and IRF-7 were up-regulated in and released from granulocytes and macrophages/dendritic cells. However, type I IFN was dispensable in the development of acute lung inflammation and pulmonary fibrosis induced by silica exposure (21). We found that the IFN-β precursor was up-regulated in CB, compared with SI and SII (Table 1; Figure 2), suggesting cytokine was important in CB but not silicosis.
Interleukin 6 (IL-6) acted as a pro-inflammatory cytokine and anti-inflammatory myokine, and IL-6 secreted by T-cells and macrophages stimulated the immune response and played a role in fighting infection. Its role as an anti-inflammatory cytokine was mediated via its inhibitory effects on TNF-α and IL-1. IL-6 was also responsible for stimulating acute phase protein synthesis and supporting the growth of B-cells. Increased inflammatory markers included IL-6 and IL-8, TNF-alpha and C-reactive protein (CRP) that participated in the pulmonary inflammatory cascade (22). High expression of CRP, IL-6, IL-8 and IL-10 were associated with diminished forced expiratory volume and increased dyspnea, and diagnostic for chronic obstructive lung disease stages 3 and 4 (23). The Th2 cytokine network in the lung was reportedly mediated by the IL-6 receptor in silica-induced pulmonary fibrosis (24). IL-6 and IL-12 receptor β1 isoform CRA_c were both up-regulated in silicosis serum compared with CB patients (Figure 1, Table 1), which implied that IL-6 may perform an essential role in silicosis but a different, lesser role in CB.
Tumor necrosis factor (TNF) was a pro-inflammatory cytokine involved in the innate immune response and had been implicated in tumor regression. The TNF protein was synthesized as a pro-hormone with a long and atypical signal sequence and the mature processed form was secreted after cleavage of the pro-peptide (25). TNF was essential in apoptosis, tumor cell death, cell proliferation and differentiation. TNK, TNF receptor 13B variant and TNF ligand superfamily member 4 isoform X1 were all up-regulated in silicosis compared to CB (Table 1; Figures 1,2). Exposure of AMs to coal dust particles triggered a significant release of TNF with titanium dioxide as a biologically inert control dust (8,11). TNF played an important role in inflammation and development of silica-induced pulmonary fibrosis, and a marked and persist increase in TNF mRNA in lung was observed after intratracheal inhalation of silica (26). Additionally, silica-induced collagen production was reduced by anti-TNF antibodies (26).
Glycoproteins and apolipoproteins
Alpha-1-B glycoprotein (A1BG) was a plasma glycoprotein encoded by the A1BG gene, and the A1BG polypeptide had four N-linked glucosamine oligosaccharides. A1BG appeared to have evolved from an ancestral gene similar to the immunoglobulin supergene family (27). However, the biological function of A1BG was not elucidated, but its expression was elevated in several cancers including cervical intraepithelial neoplasia (28), which suggested it may be play a role in carcinogenesis. A1BG was up-regulated in CB compared to silicosis (Table 1; Figures 1,2), suggesting a possible involvement in the immune reaction in CB.
Apolipoproteins (Apos) bind lipids to form lipoproteins for transport of lipids through the lymphatic and circulatory systems. Apos acted as structural components of lipoprotein particles, as cofactors for enzymes and ligands for cell-surface receptors in lipid transport that regulated the metabolism of lipoproteins and their uptake. The gene encoding ApoA-IV contained three exons separated by two introns and the primary translation product was a 396 residue pre-protein. Apo A-IV was a potent activator of lecithin-cholesterol acyltransferase in vitro, improved glucose homeostasis by enhancing pancreatic insulin secretion in the presence of elevated levels of glucose, and also reduced hepatic gluconeogenesis via nuclear receptor NR1D1 (29).
Turpentine oil injection experiments on an animal model showed that downregulation of ApoA-I and ApoA-IV transcripts in the liver may be mediated by IL-6 and TNF-α (30). Up-regulation of the ApoA-IV precursor in CB relative to SI and SII (Table 1, Figure 1) indicated a possible involvement in lipoprotein transport in the pathophysiology of CB.
Keratins
Keratins formed a family of fibrous structural proteins consisting of two types of intermediate filament that made up one of the major structural fibers of epithelial cells. Keratin intermediate filaments consisted of copolymers of specific type I and type II keratin proteins involved in the etiology of a large heterogeneous group of hereditary epithelial diseases (31). We found that keratin type II cuticular Hb4 was up-regulated in silicosis (Table 1; Figure 1). The possible connection to the etiology of pulmonary disease was not clear, although keratin K6 was previously detected in silica dust-induced trans-differentiated lung fibroblasts in rats (32). Keratins maybe therefore participate in fibrosis during silicosis.
Immune system
HLA class II histocompatibility antigen DRB1-9 beta chain, encoded by HLA-DRB1 DRB1, was the most prevalent β-subunit of HLA-DR that played a central role in the immune system by presenting peptides derived from extracellular proteins, and was associated with rheumatoid arthritis. The frequency of HLA-DRB1*0406 was significantly higher in Caspase-8-positive silicosis patients (16.67%) than in control individuals (3.03%, P=0.0006) (33), and HLA-DRB1 had been associated with rheumatoid arthritis-related pulmonary fibrosis (34). Up-regulation of HLA-DRB1 in silicosis compared to CB patients (Figures 1,2; Table 1) further supported the role of HLA-DRB1 in the development of silicosis and indicated a different role in CB.
Oncogene-related proteins
C-fos, a proto-oncogene member of the Fos family of transcription factors, has been mapped to chromosome region 14q21→q31 and formed a heterodimer with c-jun. Together, these proteins constitute the Activator Protein-1 (AP-1) complex that bound DNA at AP-1-specific sites to stimulate transcription of AP-1 responsive genes in response to extracellular signals (35). AP-1 complex binding to promoter regions may govern inflammation, proliferation and apoptosis, since Fos proteins were known to be involved in regulating cell proliferation, differentiation and transformation, and in some cases, apoptotic cell death (36). Initiation of fibroblast proliferation induced by silica dust may emerge after upregulation of the early response protooncogenes c-fos and c-jun (37). Compared with CB, c-Fos was up-regulated in silicosis patients (Figures 1,2; Table 1), suggesting it may be specifically involved in the pathology of silicosis.
Other categories
Medullasin was an inflammatory serine protease of bone marrow cells that modified the functions of natural killer (NK) cells, monocytes and granulocytes. Medullasin in granulocytes appeared to play an essential role in the development of inflammation involved in bio-defense mechanism. Medullasin enhanced the activity of NK cells by stimulating the maturation of large granular lymphocytes into NK cells in human lymphocytes (38), and it may also affect metabolism by modulating cytokine and growth factors following activation of macrophages (39). However, the postulated role of medullasin in collagen metabolism during excessive growth in gingival tissue was not confirmed (40). Medullasin was slightly upregulated in silicosis compared with CB patients (Figures 1,2; Table 1), indicating a possible role in the regulation of collagen metabolism and immune function in silicosis.
Conclusions
In order to investigate differences in silicosis and CB, proteomic profiling was performed on serum from silicosis (stage I and II) and CB patients by using 2D-gel electrophoresis and mass spectrometry. A number of proteins were found to be significantly up-regulated in silicosis patients (Table 1; Figure 1), including interferons, interleukins and TNF cytokines. By contrast, transforming growth factor beta-1 precursor and interferon beta precursor were markedly up-regulated in CB. IL-6, TNF and TNF receptor 13B variant were also up-regulated in silicosis, whereas glycoprotein- and apolipoprotein-associated proteins ApoA-IV and α-1-B-glycoprotein were more abundant in CB serum, suggesting the immune response and lipoprotein transport may be involved in the pathogenesis of CB. HLA-DRB1, medullasin and proto-oncogene c-Fos were also upregulated in silicosis, indicating different roles for these proteins in these two diseases. These proteomic profiling results provided new insight into the possible pathological mechanisms involved in disease development and progression, and would facilitate future research on the diagnosis and potential therapeutic targets of these two debilitating pulmonary diseases.
Acknowledgements
Funding: This work was supported by the Jiangsu Provincial Special Foundation for People’s Livelihoods of Science and Technology [Grant No. BL2014024] and the Jiangsu Provincial Foundation for Six Talent Summit [Grant No. 2014WSN063].
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Kim V, Criner GJ. Chronic bronchitis and chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2013;187:228-37. [Crossref] [PubMed]
- Hargreave FE, Parameswaran K. Asthma, COPD and bronchitis are just components of airway disease. Eur Respir J 2006;28:264-7. [Crossref] [PubMed]
- Kim V, Han MK, Vance GB, et al. The chronic bronchitic phenotype of COPD: an analysis of the COPDGene Study. Chest 2011;140:626-33. [Crossref] [PubMed]
- Hogg JC, Chu F, Utokaparch S, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med 2004;350:2645-53. [Crossref] [PubMed]
- Zhu Z, Homer RJ, Wang Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest 1999;103:779-88. [Crossref] [PubMed]
- Kikly K, Liu L, Na S, et al. The IL-23/Th(17) axis: therapeutic targets for autoimmune inflammation. Curr Opin Immunol 2006;18:670-5. [Crossref] [PubMed]
- Driscoll KE, Lindenschmidt RC, Maurer JK, et al. Pulmonary response to silica or titanium dioxide: inflammatory cells, alveolar macrophage-derived cytokines, and histopathology. Am J Respir Cell Mol Biol 1990;2:381-90. [Crossref] [PubMed]
- Vanhée D, Gosset P, Boitelle A, et al. Cytokines and cytokine network in silicosis and coal workers' pneumoconiosis. Eur Respir J 1995;8:834-42. [PubMed]
- Wallaert B, Lassalle P, Fortin F, et al. Superoxide anion generation by alveolar inflammatory cells in simple pneumoconiosis and in progressive massive fibrosis of nonsmoking coal workers. Am Rev Respir Dis 1990;141:129-33. [Crossref] [PubMed]
- Rolfe MW, Kunkel SL, Standiford TJ, et al. Pulmonary fibroblast expression of interleukin-8: a model for alveolar macrophage-derived cytokine networking. Am J Respir Cell Mol Biol 1991;5:493-501. [Crossref] [PubMed]
- Gosset P, Lassalle P, Vanhée D, et al. Production of tumor necrosis factor-alpha and interleukin-6 by human alveolar macrophages exposed in vitro to coal mine dust. Am J Respir Cell Mol Biol 1991;5:431-6. [Crossref] [PubMed]
- Postlethwaite AE, Seyer JM. Stimulation of fibroblast chemotaxis by human recombinant tumor necrosis factor alpha (TNF-alpha) and a synthetic TNF-alpha 31-68 peptide. J Exp Med 1990;172:1749-56. [Crossref] [PubMed]
- Dinarello CA. Inflammatory cytokines: interleukin-1 and tumor necrosis factor as effector molecules in autoimmune diseases. Curr Opin Immunol 1991;3:941-8. [Crossref] [PubMed]
- LeRoith D, McGuinness M, Shemer J, et al. Insulin-like growth factors. Biol Signals 1992;1:173-81. [Crossref] [PubMed]
- Bornstein P, Sage H. Regulation of collagen gene expression. Prog Nucleic Acid Res Mol Biol 1989;37:67-106. [Crossref] [PubMed]
- Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998;95:14863-8. [Crossref] [PubMed]
- Conesa A, Götz S, García-Gómez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005;21:3674-6. [Crossref] [PubMed]
- Sakai K, Jawaid S, Sasaki T, et al. Transforming growth factor-β-independent role of connective tissue growth factor in the development of liver fibrosis. Am J Pathol 2014;184:2611-7. [Crossref] [PubMed]
- Li L, Ruan Y, Chen Y, et al. The role of transforming growth factor-beta(1) in smoking-induced chronic bronchitis and emphysema in hamsters. Zhonghua Jie He He Hu Xi Za Zhi 2002;25:284-6. [PubMed]
- George PM, Oliver E, Dorfmuller P, et al. Evidence for the involvement of type I interferon in pulmonary arterial hypertension. Circ Res 2014;114:677-88. [Crossref] [PubMed]
- Giordano G, van den Brûle S, Lo Re S, et al. Type I interferon signaling contributes to chronic inflammation in a murine model of silicosis. Toxicol Sci 2010;116:682-92. [Crossref] [PubMed]
- Barnes PJ, Celli BR. Systemic manifestations and comorbidities of COPD. Eur Respir J 2009;33:1165-85. [Crossref] [PubMed]
- Piehl-Aulin K, Jones I, Lindvall B, et al. Increased serum inflammatory markers in the absence of clinical and skeletal muscle inflammation in patients with chronic obstructive pulmonary disease. Respiration 2009;78:191-6. [Crossref] [PubMed]
- Tripathi SS, Mishra V, Shukla M, et al. IL-6 receptor-mediated lung Th2 cytokine networking in silica-induced pulmonary fibrosis. Arch Toxicol 2010;84:947-55. [Crossref] [PubMed]
- Sherry B, Jue DM, Zentella A, et al. Characterization of high molecular weight glycosylated forms of murine tumor necrosis factor. Biochem Biophys Res Commun 1990;173:1072-8. [Crossref] [PubMed]
- Piguet PF, Collart MA, Grau GE, et al. Requirement of tumour necrosis factor for development of silica-induced pulmonary fibrosis. Nature 1990;344:245-7. [Crossref] [PubMed]
- Ishioka N, Takahashi N, Putnam FW. Amino acid sequence of human plasma alpha 1B-glycoprotein: homology to the immunoglobulin supergene family. Proc Natl Acad Sci U S A 1986;83:2363-7. [Crossref] [PubMed]
- Canales NA, Marina VM, Castro JS, et al. A1BG and C3 are overexpressed in patients with cervical intraepithelial neoplasia III. Oncol Lett 2014;8:939-47. [PubMed]
- Li X, Xu M, Wang F, et al. Apolipoprotein A-IV reduces hepatic gluconeogenesis through nuclear receptor NR1D1. J Biol Chem 2014;289:2396-404. [Crossref] [PubMed]
- Navarro MA, Carpintero R, Acín S, et al. Immune-regulation of the apolipoprotein A-I/C-III/A-IV gene cluster in experimental inflammation. Cytokine 2005;31:52-63. [Crossref] [PubMed]
- Smith F. The molecular genetics of keratin disorders. Am J Clin Dermatol 2003;4:347-64. [Crossref] [PubMed]
- Hao CF, Li XF, Yao W. Protein expression in silica dust-induced transdifferentiated rats lung fibroblasts. Biomed Environ Sci 2013;26:750-8. [PubMed]
- Ueki A, Isozaki Y, Kusaka M. Anti-caspase-8 autoantibody response in silicosis patients is associated with HLA-DRB1, DQB1 and DPB1 alleles. J Occup Health 2005;47:61-7. [Crossref] [PubMed]
- Ennis H, Gupta A, Dawson J, et al. HLA-DRB1 associations with rheumatoid arthritis-related pulmonary fibrosis. Scand J Rheumatol 2014;43:75-6. [Crossref] [PubMed]
- Chiu R, Boyle WJ, Meek J, et al. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 1988;54:541-52. [Crossref] [PubMed]
- Watanabe T, Hiasa Y, Tokumoto Y, et al. Protein kinase R modulates c-Fos and c-Jun signaling to promote proliferation of hepatocellular carcinoma with hepatitis C virus infection. PLoS One 2013;8:e67750. [Crossref] [PubMed]
- Janssen YM, Heintz NH, Marsh JP, et al. Induction of c-fos and c-jun proto-oncogenes in target cells of the lung and pleura by carcinogenic fibers. Am J Respir Cell Mol Biol 1994;11:522-30. [Crossref] [PubMed]
- Aoki Y, Sumiya M, Oshimi K. Medullasin enhances human natural killer cell activity. J Clin Invest 1982;69:1223-30. [Crossref] [PubMed]
- Havemose-Poulsen A, Holmstrup P. Factors affecting IL-1-mediated collagen metabolism by fibroblasts and the pathogenesis of periodontal disease: a review of the literature. Crit Rev Oral Biol Med 1997;8:217-36. [Crossref] [PubMed]
- Ozaki Y, Kunimatsu K, Hara Y, et al. An involvement of granulocyte medullasin in phenytoin-induced gingival overgrowth in rats. Jpn J Pharmacol 2002;89:235-41. [Crossref] [PubMed]


