Plasma microRNAs expression profile in female workers occupationally exposed to mercury
Introduction
Mercury (Hg) is a heavy metallic element that exists widely in nature (1). There are three forms of mercury: elemental, inorganic and organic. Elemental mercury is the main form of mercury generally used in industry (1). Exposure to elemental mercury of the general population and in occupational settings is primarily through inhalation of its vapor (2). Occupational exposure to mercury basically occurs in mercury thermometer plants, fluorescent light bulb manufacturing factories and small-scale gold mining (3).
Although in October 2013, China has signed the Minamata Convention on Mercury to ban the use of mercury-containing thermometers before 2020 (4,5). The problem of occupational mercury poisoning still exists in plants involved in the manufacture of mercury-containing thermometer (6). Leakage during the production process of mercury thermometer may expose workers to mercury vapor (6). In human, mercury vapor (Hg0) is readily absorbed through the respiratory tract (~80%) (7). Mercury vapor is fat-soluble and can be absorbed through alveolar epithelial cells (absorptivity can up to 70%) (8). Mercury is initially concentrated in liver and then it is transported to kidney (9). Also, it can pass blood-brain barrier and accumulate in brain tissues for a long time (10,11). Mercury mainly induces damages of nervous system (10), kidney (9), and cardiovascular system (inducing high blood pressure, coronary heart disease, myocardial infarction, arrhythmia and atherosclerosis) (12). Several studies have found that high prenatal exposure to mercury of female in reproductive period has been related with increased preterm birth risk (13) and other adverse birth outcomes (14).
MicroRNAs (miRNAs) are a family of endogenously expressed small single stranded regulatory RNAs that play critical roles in regulation of gene expression by pairing to the target mRNAs or inhibiting the translation of mRNA (15). The expressions of miR-204, miR-211, miR-448, miR-449a, miR-34b, and miR-34c may be associated with neurophysiological pathways and neurodegenerative diseases in rats following chronic lead exposure (16). Another research from Hong Kong showed that miR-21 may involve in the pathogenetic mechanisms linking heavy metals (arsenic and lead) exposure and albuminuria (17). Several studies have explored the miRNAs expression profiles of general population environmentally exposed to mercury. Sanders et al. (18) assessed the association between miRNA expression in the cervix with mercury levels during pregnancy and found 17 miRNAs were negatively associated with toenail mercury levels. However, few studies have investigated potential effects of occupational mercury exposure on miRNA expression profiles in plasma.
Plasma miRNAs have the potential to serve as stable blood-based biomarkers of physiological and pathological conditions (19). The current study was designed to identify plasma miRNAs profiles that were associated with mercury exposure by using microarray chip an effective approach for the high-throughput analysis of miRNAs expression profile, and subsequent validation by real-time PCR.
Methods
Study subjects
This study was approved by the Ethics Committee of Jiangsu Provincial Center for Disease Control and Prevention (Nanjing, China). Written informed consent was obtained from all the subjects who participated in the study.
All of our subjects were genetically unrelated ethnic Han Chinese and came from one mercury thermometer factory in Eastern China. When the employees of that factory had finished their occupational health examination in 2013, we enrolled 237 workers (20 male workers and 217 female workers) who were employed at the mercury thermometer plants for at least 1 year and worked in various operation posts including scrubbing mercury, injecting mercury, vacuumizing, constricting, maintenance, separating, fix-pointing, encapsulating, printing and examination (all exposed to mercury); or in offices. We excluded (I) male workers because men made up less than 10% of all the workers; (II) workers with a self-reported history of chronic diseases, including cancers, primary renal disease; workers who have taken any medicines in the preceding 3 months; and (III) workers who did not provide urine and/or blood samples. Thus, a total of 126 workers participated in all the parts of this study. After participants provided informed consent, we administered a questionnaire to collect information on demographic characteristics, smoking and drinking habits, medical history, drug history and occupational experiences. Participants were not considered smokers unless they had smoked an average of <1 cigarette/day for <1 year in their lifetime (nonsmokers), and not considered drinkers unless they had drunk alcoholic beverages less than once each week for <1 year in their lifetime (nondrinkers). Then the participants attended an occupational health examination, including urine mercury measurement, whole blood collection and physical examination. The urine mercury measurement was based on Urine-Determination of mercury-Cold atomic absorption spectrometric method-II Acidic stannous chloride reduction method (WS/T26-1996). The plasma was isolated from a fresh individual whole blood sample in BD Vacutainer® Venous Blood Collection Tubes (cat. no. 367525) containing ethylene diamine tetraacetic acid (EDTA) within 4 hours. All plasma samples were stored at −80 °C.
Study design
To identify mercury-related plasma miRNAs in workers, a case-control study was designed to analyze the plasma miRNA profile between (I) chronic mercury poisoning group; (II) mercury absorbing group and (III) control group in a mercury thermometer plant. We clearly defined chronic mercury poisoning group, mercury absorbing group and control group by Diagnostic Criteria of Occupational Mercury Poisoning (GBZ 89-2007) of China. The normal reference value of urine mercury (Hg-U) was less than 4 µg/g Cr for normal person in China. Increased urine mercury was defined as over 35 µg/g Cr in workers occupationally exposed to mercury. Clinical symptoms of chronic mercury poisoning includes: (I) neurasthenic syndrome; (II) stomatitis—gingivitis; (III) tremor and; (IV) proximal renal tubular dysfunction. Thus, in our research, we selected people working in the offices with urine mercury less than 4 µg/g Cr as control group. Mercury absorbing group was defined as workers with increased urine mercury but not clinical symptoms of chronic mercury poisoning. And chronic mercury poisoning group was defined as workers who had any three of the following symptoms: (I) neurasthenic syndrome; (II) stomatitis—gingivitis; (III) tremor; (IV) proximal renal tubular dysfunction and (V) increased urine mercury.
We firstly selected ten chronic mercury poisoning patients. Mercury absorbing individuals and control group individuals were matched to these patients one by one. The general matching principles of all the subjects were age (±3 years), sex (female), nationality (“Han”), smoking (nonsmokers) and drinking (nondrinkers) status to minimize their confounding effects on miRNA expression profile. We prepared a 1.5-mL pooled plasma sample for each group which included 150 µL of plasma from each subject. We then subjected three plasma pools to miRNA microarray assay and compared miRNA expression profiles between these three groups. To focus on the most likely related miRNAs in the validation stage, miRNAs were selected based on the following criteria: (I) miRNA expressions are up-regulated or down-regulated in the mercury poisoned group compared to the other two groups; (II) demonstrated at least a 1.5-fold lower or higher expression between three groups; (III) expressed at least 50 copies in all the groups; and (IV) giving priority to choosing the miRNA that may be associated with nervous, cardiovascular system and kidney damage based on an extensive literature review. The plasma samples prepared for the validation stage were identical to the samples for microarray analysis.
Purification of total RNA
In preparation for microarray analysis, three plasma pools were produced by the mixed ten samples per group (150 µL per sample) for the microfluidic microarray analysis. The pooled plasma was then subjected to the total RNA extraction using miRNeasy Serum/Plasma Kit (QIAGEN, cat. no. 217184, Germany). In the validation stage, RNAs were extracted from individual plasma samples (200 µL per sample) using a miRNeasy Serum/Plasma Kit (QIAGEN, cat. no. 217184, Germany). A synthetic Caenorhabditis elegans miRNA [cel-miR-238 (20 fmol); Takara, Dalian, China] was added to each plasma sample as an internal control prior to the isolation procedure.
MicroRNA microarray assay
The microarray assay began from 4 to 8 µg of the purified total RNA samples and was labeled with a 3’-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for subsequent fluorescent dye staining. Hybridization was then performed overnight on a µParaflo® microfluidic microarray using a micro-circulation pump (Atactic Technologies). On the microfluidic microarray, each detection probe consisted of a chemically modified nucleotide coding segment complementary to either the target microRNA (from miRBase, http://www.mirbase.org/) or the other RNA (control or customer defined sequences) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. Following RNA hybridization, tag-conjugating Cy3 dyes were circulated through the microfluidic microarray for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Device). Finally, the fluorescence images were converted into digital-quality data using Array-Pro image analysis software (Media Cybernetics). The data were analyzed by firstly subtracting the background and then normalizing the signals which used a LOWESS filter (locally weighted regression) (20).
qRT-PCR validation
The expression levels of the candidate miRNAs selected from the microarray analysis were validated using TaqMan miRNA qRT-PCR primers and reagents. Purified total RNA (1.67 µL) from the individual plasma sample was subjected to reverse transcription in a 5-µL reaction mixture using miRNA-specific stem-loop primers and a TaqMan® miRNA Reverse Transcription Kit (Applied Biosystems, USA) following manufacturer’s protocol. The real-time PCR assay was performed in a 10-µL reaction mixture that contained 5 µL of 2× TaqMan Universal PCR Master Mix (No AmpErase UNG), 4.5 µL of diluted cDNA (1:15) and 0.5 µL of TaqMan miRNA Assay primers (Applied Biosystems, USA). The thermal cycling procedure was set as follows: an initial denaturation step at 95 °C for 10 min, 40 cycles of PCR amplification at 95 °C for 15 s, and 60 °C for 1 min. Each extracted total RNA sample for each candidate miRNA was run in triplicate. The expression level of each miRNA was individually normalized to cel-miR-238.
Data analysis
The relative expression level of each candidate miRNA was individually calculated by 2-deltaCt, of which delta cycle threshold (ΔCt) = Ct miRNA−Ct cel-miR-238. The relative quantification value then underwent a log2-transformation to compare the expression levels of the candidate miRNAs between groups. The statistical analysis was performed by using SPSS 21.0 software, and a P value <0.05 was considered as statistically significant.
Results
Subject characteristics
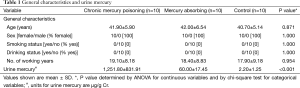
As shown in Table 1, the distribution of age, sex, smoking status, drinking status and working years were matched between (I) chronic mercury poisoning group; (II) mercury absorbing group and (III) control group (all P>0.05), whereas the urine mercury levels were all significantly different between three groups (all P≤0.001).
Full table
miRNA expression profiles and miRNA selection for validation
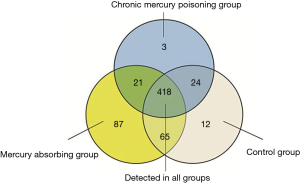
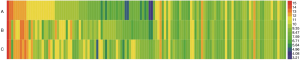
The expression levels of 2,557 human miRNAs listed in Sanger miRBase Release 20.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/) were tested based on the µParaflo® microfluidic microarray. As shown in Figure 1, 630 miRNAs were identified in at least one group and 418 miRNAs were detected in all the three groups. Figure 2 showed the heat map of 128 miRNAs, the expressions of which were over 50 copies in all three groups.
Based on the miRNA selection criteria (see “Materials and Methods”), we selected eight miRNAs, the expressions of which were up-regulated or down-regulated in the mercury poisoning group compared to the other two groups. There were four miRNAs (miR-16-5p, miR-30c-3p, miR-181a-5p and let-7e-5p) showing down-regulation and four miRNAs (miR-92a-3p, miR-122-5p, miR-451a and miR-486-5p) showing up-regulation. The expression levels and related functions of eight selected miRNAs are shown in Table 2.

Full table
Identification of mercury-related miRNAs
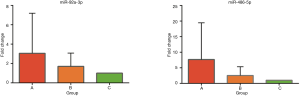
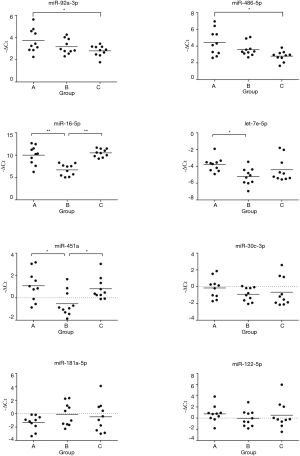
In the validation stage, eight miRNAs (miR-16-5p, miR-30c-3p, miR-181a-5p, let-7e-5p, miR-92a-3p, miR-122-5p, miR-451a and miR-486-5p) were selected for the TaqMan Assay by qRT-PCR in three groups. The plasma samples were identical to the samples used in the microarray stage. The plasma levels of the eight miRNAs are displayed in Figure 3. The relative levels (-ΔCt) of plasma miR-92a-3p were 3.72±1.06, 3.18±0.67 and 2.81±0.49 (mean ± SD) in mercury poisoning group, mercury absorbing group and control group respectively. The relative levels (-ΔCt) of plasma miR-486-5p were 4.44±1.53, 3.59±0.79 and 2.80±0.61 (mean ± SD) in mercury poisoning group, mercury absorbing group and control group respectively. As shown in Figure 3, the expression levels of miR-92a-3p and miR-486-5p were significantly up-regulated (P<0.05) in the mercury poisoning group compared to the other two groups. The results were consistent with the microarray analysis. The fold changes of miR-92a-3p and miR-486-5p are shown in Figure 4.
Discussion
So far, the toxicity mechanism of mercury poisoning is mainly about its high-affinity with sulfhydryl groups (12,29,30). Molecular interactions with sulfhydryl groups in molecules of albumin, metallothionein, glutathione, and cysteine have been implicated in mechanisms involved in the proximal tubular uptake, accumulation, transport, and toxicity of mercuric ions (29). However, it can’t fully explain why mercury had toxic effects on a number of organs, especially in the cardiovascular systems.
A lot of studies suggested that environmental metal pollutants can induce changes in miRNA expression (16-18). In our research, miRNA microarray assay and subsequent validation revealed that miR-92a and miR-486 were positively associated with mercury exposure. MiR-92a and miR-486 might prove to be potential biomarkers for mercury exposure.
A recent research found that, miR-92a was significantly up-regulated in neointimal lesions after wire-induced injury and inhibition of endothelial miR-92a attenuates neointimal lesion formation through the acceleration of re-endothelialization and thus represents a putative novel mechanism to enhance functional recovery following the injury vascular (31). Another study also showed that miR-92a was up-regulated by oxLDL in atheroprone areas which promoted endothelial activation and the development of atherosclerotic lesions (25). Thus, the up-regulation of miR-92a in mercury exposed workers implied that miR-92a may have associations with the endothelial dysfunction and the formation of atherosclerotic lesions in mercury poisoning.
The study on the function of miR-486 reveals that miR-486 can directly suppress nuclear factor-κB (NF-κB)-negative regulators, CYLD and Cezanne, as well as multiple A20 activity regulators, resulted in promotion of ubiquitin conjugations in NF-κB signaling and sustained NF-κB activity (28). Moreover, up-regulation of miR-486 promotes glioma aggressiveness through activation of NF-κB signaling pathway both in vitro and in vivo study (28). Importantly, miR-486 levels in primary gliomas significantly correlated with NF-κB activation status (28). A previous investigation found that mercury alone can induce NF-κB activation, result in the induced expression of COX-2 and iNOS which can induce inflammatory diseases (32). Thus, the up-regulation of miR-486 which can enhance NF-κB activation may be associated with mercury poisoning.
Plasma miR-16, miR-451a and let-7e expressions of group B are down-regulated comparing to the other two groups. Although lacking direct supporting evidence, we attempted to further comprehend the miRNA expression pattern of this group. Normally, miRNAs were selectively packaged into microvesicles (MVs) in cells and actively secreted out of the cells (33). Plasma miRNAs are generated from different tissues and organs which have different sensitivity to the mercury. Thus, mercury poisoning may cause cell necrosis or apoptosis of different cells. When cells had necrosis, miRNAs were directly released from the cells (34) and causing the up-regulations of these down-regulated miRNAs (miR-16, miR-451a and let-7e of group B) in group A. This may help to explain the miRNA expression pattern of group B, but further research is needed.
Our study also has several limitations. Firstly, our study subject is a sexual homogeneous population. Thus, our results may likely be better generalized to female population and limit external generalizability. Secondly, we sampled only 30 individuals and thus whether miR-92a and miR-486 are potential circulating biomarkers should be validated in studies with large samples. Thirdly, data were incomplete, we are unable to analyze the relationship between miRNAs expression and hair or nail Hg (chronic biomarkers of biological levels of Hg). Additionally, although samples were matched, we are unable to adjust for covariates. As our research is an epidemiological study, it is difficult to determine whether the differences in miRNAs expression preceded or followed Hg exposure and whether the miRNAs are results of physiological damage which has occurred due to Hg exposure. More mechanistic studies of miR-92a and miR-486 in mediating toxicity should be validated in cell or animal models in future studies.
Conclusions
MiR-92a and miR-486 might prove to be potential biomarkers for mercury exposure. The up-regulation of miR-92a and miR-486 of mercury exposed female workers may have associations with the endothelial dysfunction which caused the formation of atherosclerotic lesions, the enhancement of NF-κB activation which induced inflammatory diseases respectively. Studies have demonstrated that reproductive tissues are susceptible to environmental contaminants (35) and high prenatal exposure to mercury of female in reproductive period has been related with numerous adverse birth outcomes (14). Some of these female workers have worked for many years in the plant and are occupationally exposed to high levels of Hg during their reproductive years. There are probably reproductive issues for these women and this represents a public health concern which is a hot spot of future research. Furthermore, further studies are necessary to prove the causal association between miRNAs changes and mercury exposure, and determine whether these two miRNAs are clear biomarkers to mercury exposure.
Acknowledgements
Funding: This work was supported by Outstanding Medical Academic Leaders program of Jiangsu Province (LJ201130).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Tchounwou PB, Ayensu WK, Ninashvili N, et al. Environmental exposure to mercury and its toxicopathologic implications for public health. Environ Toxicol 2003;18:149-75. [Crossref] [PubMed]
- Risher JF, Murray HE, Prince GR. Organic mercury compounds: human exposure and its relevance to public health. Toxicol Ind Health 2002;18:109-60. [Crossref] [PubMed]
- Risher JF, Nickle RA, Amler SN. Elemental mercury poisoning in occupational and residential settings. Int J Hyg Environ Health 2003;206:371-9. [Crossref] [PubMed]
- Kirby T. UN agrees new treaty to reduce harm from mercury. Lancet 2013;381:362. [Crossref] [PubMed]
- Larson HJ. The Minamata Convention on Mercury: risk in perspective. Lancet 2014;383:198-9. [Crossref] [PubMed]
- Tang N, Li YM. Neurotoxic effects in workers of the clinical thermometer manufacture plant. Int J Occup Med Environ Health 2006;19:198-202. [Crossref] [PubMed]
- Holmes P, James KA, Levy LS. Is low-level environmental mercury exposure of concern to human health? Sci Total Environ 2009;408:171-82. [Crossref] [PubMed]
- Reichl FX, Walther UI, Durner J, et al. Cytotoxicity of dental composite components and mercury compounds in lung cells. Dent Mater 2001;17:95-101. [Crossref] [PubMed]
- Pelclová D, Lukás E, Urban P, et al. Mercury intoxication from skin ointment containing mercuric ammonium chloride. Int Arch Occup Environ Health 2002;75 Suppl:S54-9. [Crossref] [PubMed]
- Langworth S, Almkvist O, Söderman E, et al. Effects of occupational exposure to mercury vapour on the central nervous system. Br J Ind Med 1992;49:545-55. [PubMed]
- Eto K, Takahashi H, Kakita A, et al. Pathological and biochemical studies of 30 Niigata autopsy cases related to Minamata disease. Nihon Eiseigaku Zasshi 2007;62:70-88. [PubMed]
- Houston MC. Role of mercury toxicity in hypertension, cardiovascular disease, and stroke. J Clin Hypertens (Greenwich) 2011;13:621-7. [Crossref] [PubMed]
- Xue F, Holzman C, Rahbar MH, et al. Maternal fish consumption, mercury levels, and risk of preterm delivery. Environ Health Perspect 2007;115:42-7. [Crossref] [PubMed]
- Silbergeld EK, Patrick TE. Environmental exposures, toxicologic mechanisms, and adverse pregnancy outcomes. Am J Obstet Gynecol 2005;192:S11-21. [Crossref] [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [Crossref] [PubMed]
- An J, Cai T, Che H, et al. The changes of miRNA expression in rat hippocampus following chronic lead exposure. Toxicol Lett 2014;229:158-66. [Crossref] [PubMed]
- Kong AP, Xiao K, Choi KC, et al. Associations between microRNA (miR-21, 126, 155 and 221), albuminuria and heavy metals in Hong Kong Chinese adolescents. Clin Chim Acta 2012;413:1053-7. [Crossref] [PubMed]
- Sanders AP, Burris HH, Just AC, et al. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics 2015;7:885-96. [Crossref] [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [Crossref] [PubMed]
- Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185-93. [Crossref] [PubMed]
- Heyn J, Ledderose C, Hinske LC, et al. Adenosine A2A receptor upregulation in human PMNs is controlled by miRNA-214, miRNA-15, and miRNA-16. Shock 2012;37:156-63. [Crossref] [PubMed]
- Roy S, Benz F, Vargas Cardenas D, et al. miR-30c and miR-193 are a part of the TGF-β-dependent regulatory network controlling extracellular matrix genes in liver fibrosis. J Dig Dis 2015;16:513-24. [Crossref] [PubMed]
- Mele F, Basso C, Leoni C, et al. ERK phosphorylation and miR-181a expression modulate activation of human memory TH17 cells. Nat Commun 2015;6:6431. [Crossref] [PubMed]
- Liu J, Zhu L, Xie GL, et al. Let-7 miRNAs Modulate the Activation of NF-κB by Targeting TNFAIP3 and Are Involved in the Pathogenesis of Lupus Nephritis. PLoS One 2015;10:e0121256. [Crossref] [PubMed]
- Daniel JM, Penzkofer D, Teske R, et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc Res 2014;103:564-72. [Crossref] [PubMed]
- Du Y, Han X, Pu R, et al. Association of miRNA-122-binding site polymorphism at the interleukin-1 α gene and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Front Med 2014;8:217-26. [Crossref] [PubMed]
- Hur W, Lee JH, Kim SW, et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int J Biochem Cell Biol 2015;64:265-76. [Crossref] [PubMed]
- Song L, Lin C, Gong H, et al. miR-486 sustains NF-κB activity by disrupting multiple NF-κB-negative feedback loops. Cell Res 2013;23:274-89. [Crossref] [PubMed]
- Zalups RK. Molecular interactions with mercury in the kidney. Pharmacol Rev 2000;52:113-43. [PubMed]
- Branco V, Canário J, Holmgren A, et al. Inhibition of the thioredoxin system in the brain and liver of zebra-seabreams exposed to waterborne methylmercury. Toxicol Appl Pharmacol 2011;251:95-103. [Crossref] [PubMed]
- Loyer X, Potteaux S, Vion AC, et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res 2014;114:434-43. [Crossref] [PubMed]
- Park HJ, Youn HS. Mercury induces the expression of cyclooxygenase-2 and inducible nitric oxide synthase. Toxicol Ind Health 2013;29:169-74. [Crossref] [PubMed]
- Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010;39:133-44. [Crossref] [PubMed]
- Yamaura Y, Nakajima M, Tatsumi N, et al. Changes in the expression of miRNAs at the pericentral and periportal regions of the rat liver in response to hepatocellular injury: comparison with the changes in the expression of plasma miRNAs. Toxicology 2014;322:89-98. [Crossref] [PubMed]
- Leino O, Kiviranta H, Karjalainen AK, et al. Pollutant concentrations in placenta. Food Chem Toxicol 2013;54:59-69. [Crossref] [PubMed]