Positive expression of protein chromosome 9 open reading frame 86 (C9orf86) correlated with poor prognosis in non-small cell lung cancer patients
Introduction
Lung cancer accounted for 13% (1.6 million) of the total cancer cases and 18% (1.4 million) of the cancer-related deaths in 2008 in the US (1). In China, the number of lung cancer cases and related-deaths has increased with increasing cigarette smoking and environmental pollution (2). Performance status, disease stage, age, sex, and amount of weight lost have been identified as survival factors of non-small cell lung cancer (NSCLC) patients (3). Surgery is the main treatment strategy for NSCLC patients with clinical stages IA to IIIA. Chemotherapy (platinum-based) after surgery may confer survival benefits in stage IB to IIIA patients (part of stage IB patients with high-risk factors) (4). Although chemotherapy improves the 5-year overall survival (OS), it is also associated with serious adverse effects. Many recent studies have focused on finding identifying novel biomarkers and new therapeutic targets in order to improve the OS, disease-free survival (DFS) and the quality of life (QOL) in patients with NSCLC.
Ras proteins are members of a large superfamily of small GTPases that have significant sequence and biochemical similarities. The Ras branch of the Ras superfamily consists of small GTPases closely related to Ras and includes the R-Ras, Rap, Ral, Rheb, Rin and Rit proteins (5). The Ras superfamily of GTPases comprises several subfamilies of small GTP-binding proteins whose functions include the control of proliferation, differentiation, and apoptosis, as well as cytoskeleton organization (6). Three decades ago, researchers found that R-Ras, similar to prototype Ras genes, may be activated as an oncogene in some human malignancies (7). Subsequently, additional Ras family proteins were identified. For example, deregulation of TC21 may trigger cellular transformation and contribute to human oncogenesis because similar signal transduction pathway is used by oncogenic Ras (8). It shows overexpression in hepatocellular carcinoma and associated with tumor progression and poor prognosis (9). The RAB5A gene is involved in the transformation from AGZY83-a to the higher metastatic cell line Anip973, indicating that overexpression of the RAB5A gene is associated with neoplasia metastasis (10). In cervical cancer cells, the RAB5A gene regulates the invasion phenotype through the integrin-mediated signaling pathway (11). Rac1 is overexpressed in breast tumor cells, and its activation is closely associated with tumor aggression (12). The expression of RAB25 promotes tumor development, while down regulation of RAB25 by RNAi transfection significantly inhibits ovarian cancer growth in vitro and in vivo xenograft models (13,14). Recently, studies indicated that Rabl3 is a novel oncogene that regulates the oncological behavior of human cancer cells. Rabl3 may be a candidate controlling tumorigenesis and a new target for anti-tumor treatment (15).
Chromosome 9 open reading frame 86 (C9orf86), also known as Rab-like protein 1 (RBEL1), is located at 9q34.3 according to the National Center for Biotechnology Information database. Functional studies have shown that C9orf86 is a novel subfamily of GTPases. C9orf86 can promote cell growth, and knockdown by RNA interference was associated with morphological and biochemical features of apoptosis as well as inhibition of extracellular signal-regulated kinase phosphorylation (16). C9orf86 can be divided into RBEL1A and RBEL1B, which localize in the nucleus and cytoplasm, respectively, and may play an important roles in breast tumorigenesis. RBEL1A was shown to be over-expressed in a large number of primary breast (67%) and colon (47%) cancer specimens compared to matching normal tissues. Knockdown of C9orf86 in MCF-7 breast cancer cells resulted in cell growth suppression associated with apoptosis and cell cycle arrest (17,18). These data indicate that C9orf86 is a potential oncogene.
The function of C9orf86 in the regulation and development of lung cancer is unclear. Therefore, in this study, we evaluated the C9orf86 expression in 180 resected lung cancer specimens and adjacent non-tumor tissues to analyze the correlation between C9orf86 protein levels and prognosis as well as clinicopathological characteristics, using immunohistochemistry (IHC) of tissue microarrays (TMAs).
Methods
Patients and tissue specimens
Paraffin-embedded, archived lung cancer specimens were obtained from 204 patients diagnosed by the Pathology Department (The First Affiliated Hospital of Guangzhou Medical University, China) from January 2002 to May 2008. In addition to the 180 informative lung cancer tissues, 16 matched adjacent noncancerous tissues were used as controls. The cases selected were based on a positive pathologic diagnosis of NSCLC in patients who underwent R0 resection without preoperative chemotherapy or radiotherapy (part of stage IV patients with single metastases and was resectable), and complete follow-up data. The NSCLC cases included 138 (76.7%) men and 42 (23.3%) women, with a mean age of 57.9 years. The average follow-up time was 45 months (median, 50 months; range, 1–111 months). Patients with unknown causes of death were excluded from our study. The clinicopathologic characteristics of the patients, including age, sex, smoking, carcinoembryonic antigen (CEA) level, lymph node metastasis, tumor size, pathologic classification, tumor size, tumor-node-metastasis (TNM) stage, lung membrane invasion, recurrence and metastasis are detailed in Table 1. We reclassified the lung adenocarcinomas (ACC) using the newest pathologic guidelines, and divide the cases into atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA) and invasive adenocarcinoma (IA) (19). Tumor stage was defined according to the American Joint Committee on Cancer (AJCC)/International Union against Cancer tumor-node-metastasis TNM classification system (20).
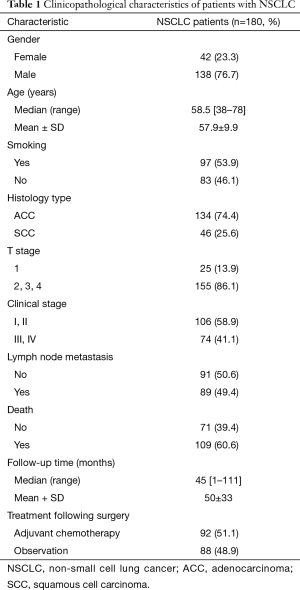
Full table
The slides were reviewed by two senior pathologists to determine and mark out representative tumor areas. Duplicates of 1-mm-diameter cylinders were punched from representative tumor areas and were used to form the tissue microarray (TMA) block (Beecher Instruments, Silver Spring, MD, USA), which contained 204 lung cancers and 16 adjacent nonmalignant lung tissues.
The experiment was approved by the Institutional Ethics Committee of Guangzhou Medical University (ID: 2015, No.21), the informed consent cannot be obtained because the specimens of lung cancer have preserved for a long time and the patients cannot be traced. An absolute confidentiality of the patients’ vital information was maintained for ethical purposes.
Cell lines and cell cultures
The NSCLC cell lines (A549, SPC-A-1, H1650, H460, H520, and H1975) and the human bronchial epithelial (16HBE) cell line were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (Gibco). All cell lines were grown in a humidified incubator 37 °C with 5% CO2.
RNA extraction and quantitative RT-PCR analysis
TRIzol reagent (Dongsheng Biotech Co., China) was used to extract the total RNA from the NSCLC cell lines (A549, SPC-A-1, H1650, H460, H520, and H1975) and 16HBE. After the reverse transcription of total RNA, the cDNA was used as a template to detect the C9orf86 expression by quantitative real-time PCR (qRT-PCR) with the SYBR Green I. The following primers were used for the PCR of C9orf86: forward 5'-CATCGTGAAGGTTGAAGTCTGG-3'; reverse 5'-GTCCACTGCTTGGTAATGTCG-3'; GAPDH forward 5'-CTGCACCACCAACTGCTTAG-3'; and GAPDH reverse 5'-AGGTCCACCACTGACACGTT-3'. The cycle threshold (Ct) values for GAPDH (reference) and C9orf86 (sample) were determined intriplicate (shown as the arithmetic mean). The quantity of C9orf86 in each NSCLC cell line relative to the average expression in 16HBE cells was calculated using the relative expression levels of C9orf86 given by 2-△△Ct, where
Western blot assay
Cells were digested in SDS lysis buffer containing 50 mM Tris-HCL (pH 7.0), 2% SDS, and 10% glycerol, and incubated for 10 min at 95 °C. Fifty micrograms of total cell lysate per lane was separated using 9% SDS-PAGE gels. A mouse mAb of C9orf86 was used for immunoblot analysis (1:700 dilution; Abnova, Taipei City, Taiwan) and mouse mAb of GAPDH (Abcam, Cambridge, UK) was used as a loading control.
Immunohistochemical staining
A commercially available antibody against C9orf86 (1:200 dilution, Mouse mAb, Abnova) was used as the primary antibody. Paraffin sections of tissues (4 µm) were deparaffinized in xylene and rehydrated in a graded alcohol series. The sections were then treated with 3% hydrogen peroxide for 10 min to quench the endogenous peroxidase activity. The antigen was retrieved by pressure cooking for 3 min in EDTA buffer (pH 8.0). The slides were then allowed to cool at room temperature and then preincubated with 10% normal goat serum at room temperature for 30 min to reduce nonspecific reactions. Subsequently, the slides were incubated with mouse monoclonal anti-C9orf86 for 2 h at room temperature. After washing with phosphate-buffered saline, the biotinylated secondary antibody was applied for 15 min at 37 °C. The sections were incubated with streptavidin–horseradish peroxidase complex and developed with 3# diaminobenzidine (DAB). Finally, the sections were counterstained with Mayer hematoxylin, dehydrated, and mounted. For negative controls, the primary antibody was replaced with normal rabbit serum. Known immunostaining positive slides were used as positive controls.
Immunohistochemical evaluation
Three investigators evaluated all specimens without knowledge of the clinical features of the cases. Variant cases were reviewed and discussed until a consensus was reached. Five areas were randomly selected and scored. The percentage of tumor cells with positive staining of C9orf86 was determined at high magnification (×200). For C9orf86, cytoplasmic immunostaining in tumor cells was considered positive. Tissue was scored (H score) based on the total percentage of positive cells and the intensity of the staining (1+, 2+, or 3+), where
The sample was considered negative if H =0 and positive if H was more than 0 (21).
Statistical analysis
All statistical analyses were performed using the SPSS 16.0 statistical software package (SPSS, Inc., Chicago, IL, USA). Because of the non-normal distribution of protein expression, statistical evaluation was performed using nonparametric tests. The χ2 test was employed to evaluate differences in expression of C9orf86 between the two categories of tissues. The χ2 test was performed to determine the correlation between C9orf86 expression and clinicopathologic characteristics. C9orf86 expression and other variables were estimated using the univariate and multivariate Cox hazards ratio model. Survival curves were plotted by Kaplan-Meier survival analysis and compared by using the log-rank test. Survival curves were further analyzed by stratifying the histology type and clinical stages. Survival time was calculated from the date of lung cancer diagnosis to the date of death from any cause. In all cases, differences with P<0.05 were considered statistically significant.
Results
C9orf86 overexpressed in NSCLC cell lines
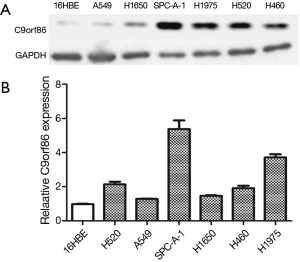
Western blotting and qRT-PCR analysis showed that C9orf86 expression in NSCLC cell lines (SPC-A-1, H460, H520, and H1975) was higher than that in 16HBE cells (Figure 1A,B).
C9orf86 overexpressed in NSCLC tissues
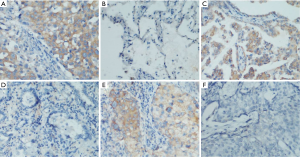
The expression of C9orf86 was investigated by IHC with a monoclonal C9orf86 antibody using a TMA containing 204 tumor tissues of NSCLC and 16 non-tumor tissues. Informative IHC data were obtained from 180 tumor tissues and 16 non-tumor tissues. C9orf86 expression was positive in the cytoplasm of tumor tissues and was not detected in non-neoplastic bronchial or alveolar epithelial cells (Figure 2A-F). Positive expression of C9orf86 in tumor tissues was observed in 74 of 180 (41.11%) NSCLC specimens (Table 2). The samples were further divided into 51 of 134 (38.06%) ACCs, 23 of 46 (50.0%) squamous cell carcinoma (SCC) and no large cell carcinomas.

Full table
Correlation between C9orf86 expression and clinicopathologic parameters
To determine whether C9orf86 expression was correlated with clinical pathological characteristics, all specimens were separated into either negative or positive C9orf86 expression groups. C9orf86 expression was significantly associated with lymph node metastasis (N stage) and clinical stage (P<0.05, Table 2) in all NSCLC patients. However, there was no significant correlation between C9orf86 expression and other clinicopathological features, such as patient age, sex, T stage, smoking, CEA level, lung membrane invasion and histology type (P>0.05, Table 2).
Positive C9orf86 expression correlated with poor survival in NSCLC patients
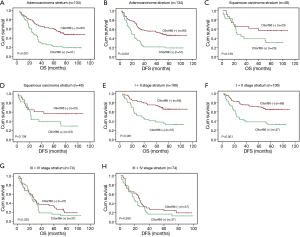
After detecting correlation between C9orf86 expression and clinicopathologic features, we examined the correlation between C9orf86 expression and patients’ survival. The 5-year OS and DFS rate of the informative 180 NSCLC patients were 41.4% and 40.4%, respectively (Figure 3A,B). Among these 180 NSCLC patients, Kaplan-Meier and log-rank test analyses indicated that C9orf86 positive expression was significantly associated with poorer OS (5-year survival rates, 27.0% vs. 55.6%, log-rank test, χ2=16.321, P<0.001, Figure 3C). Similarly, C9orf86 positive patients showed significantly shorter DFS (27.0% vs. 53.1%, log-rank test, χ2=16.159, P<0.001, Figure 3D) in all patients. To determine whether any other factors influence OS and DFS, we compared OS and DFS between the adjuvant chemotherapy and observation groups. In NSCLC patients showing C9orf86 negative expression, the OS (37.7% vs. 64.2%, log-rank test, χ2=8.922, P=0.003, Figure 3E) and DFS (34.8% vs. 63.5%, log-rank test, χ2=9.699, P=0.002, Figure 3F) in patients who received adjuvant chemotherapy were significantly prolonged. There was no significant difference between the adjuvant chemotherapy and observation groups for OS and DFS in the C9orf86 positive expression group (P>0.05, Figure 3G,H).
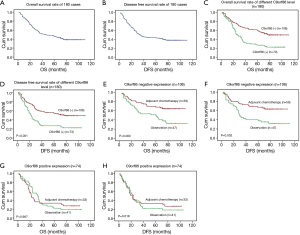
Clinical stage and histology type were taken into consideration during stratification analysis. C9orf86-negative patients showed longer OS than positive patients in the ACC stratum (21.6% vs. 51.4%, log-rank test, χ2=15.329, P<0.001, Figure 4A) and DFS (21.6% vs. 49.0%, log-rank test, χ2=15.057, P<0.001, Figure 4B). There was no significant difference between negative and positive C9orf86 expression in SCC stratum (P>0.05, Figure 4C,D). C9orf86 positive expression was significantly associated with shorter OS (37.8% vs. 66.1%, log-rank test, χ2=13.579, P<0.001, Figure 4E) and DFS (37.8% vs. 65.5%, log-rank test, χ2=12.670, P<0.001, Figure 4F) for stage I/II patients. There was no significant difference between negative and positive C9orf86 expression in III/IV stage (P>0.05, Figure 4G,H).
Univariate and multivariate analyses of prognostic variables in patients with NSCLC
To determine the clinical usefulness of using C9orf86 as a hazard ratio, we examined OS using Cox regression hazard ratio analyses. The clinicopathological factors included age, gender, smoking history, histology, T stage, N stage, clinical stage, adjuvant chemotherapy, CEA level and lung membrane invasion. As shown in Table 3, univariate Cox regression analyses revealed that positive expression of C9orf86 was associated with a significantly increased risk of cancer-related death (hazards ratio: 2.138; 95% confidence interval: 1.463–3.124; P<0.001) in NSCLC patients. C9orf86 positive expression increased the hazard ratio of lung cancer-related death by two-fold compare to C9orf86 negative expression. The hazard ratio indicated that lymph node metastasis (P<0.001), clinical stage (P<0.001), CEA level (P=0.033) and adjuvant chemotherapy (P=0.006) were worse predictors.

Full table
We adjusted for the potential confounding factors, including age, gender, smoking history, histology, T stage, N stage, clinical stage, adjuvant chemotherapy, CEA level, and lung membrane invasion. Positive expression of C9orf86 (hazards ratio: 1.823; 95% confidence interval: 1.240–2.682; P=0.002; Table 3) and clinical staging (hazards ratio: 2.532; 95% confidence interval: 1.715–3.740; P<0.001) in NSCLC were associated with poorer survival in an independent manner using a multivariate Cox regression model.
Discussion
C9orf86, also known as RBEL1, is a novel Ras superfamily protein. Previous studies showed that Ras family proteins play an important role in tumorigenesis, by promoting tumor growth and invasion and regulating processes such as cell cycle progression, proliferation, apoptosis, migration, and survival (6,7). High rates of KRAS-activating missense mutations have been detected in many cancers, including NSCLC, colon adenoma, and pancreatic ACC (22-24). Our results agree with those of previous reports showing that C9orf86 is overexpressed in primary breast cancer (17,18). In this study, qRT-PCR and western blot analysis showed that C9orf86 was highly expressed in four lung cancer cell lines. C9orf86 was also overexpressed in primary NSCLC tissues compared with their adjacent noncancerous tissues according to IHC. C9orf86 protein was expressed in 40.11% of 180 NSCLC patient specimens and was not detected in any of the 16 adjacent lung tissues.
We also demonstrated that C9orf86-positive expression was correlated with the progressive of NSCLC. C9orf86 expression was inversely correlated with clinicopathologic classification including lymph node metastasis and clinical stage (P<0.05) of NSCLC. Further analyses showed that C9orf86-positive NSCLC patients had shorter OS and DFS than C9orf86-negative patients (P<0.001). We also demonstrated a significantly increased risk of cancer-related death in patients showing positive expression of C9orf86 using univariate and multivariate Cox regression analysis. Interestingly, we found that C9orf86 with the clinical stage. Univariate Cox proportional hazard analysis revealed that N classification, clinical stage, adjuvant chemotherapy, CEA level, and C9orf86 expression were important risk factors for cancer-related death (Table 3). Furthermore, NSCLC patients with C9orf86-negative expression showed a significant survival benefit from adjuvant chemotherapy compared to patients with C9orf86-positive expression. Thus, patients with C9orf86-negative expression have early stage or no metastasis, indicating a better prognosis.
The mechanism by which C9orf86 reduces the survival time in patients with lung cancer is yet unclear. Previous reports suggest that, RhoB expression is lost in 96% of invasive tumors, and is reduced by 86% in poorly differentiated tumors compared to in non-neoplastic epithelium (25). RhoB controls apoptosis through the FKBP38-dependent regulation of Bcl-2 and Bcl-XL (26). Rab27A affects the invasive and metastatic potentials of breast cancer cells by modulating the secretion of insulin-like growth factor-II, which regulates the expression of p16, vascular endothelial growth factor, uPA, cathepsin D, cyclin D1, and matrix metalloprotease-9 (27). Rab25 overexpression has been linked to prostate cancer progression. RBEL1 promotes breast cancer cell MCF-7 and SKB-3 proliferation and inhibits apoptosis (28). These data indicate that Ras protein can promote tumor cell proliferation and invasion, serving as an oncoprotein. Based on these findings, C9orf86 may play an important role in aggression of NSCLC. Detailed investigations of phenotypic heterogeneity in NSCLC may reveal the underlying mechanism(s) to explain the role of C9orf86 in the development and progression of NSCLC. Therefore, our future studies will investigate the mechanisms underlying C9orf86-mediated progression and metastasis of NSCLC, by identifying the receptor, adapters, target proteins, and pathways of C9orf86. The expression levels of C9orf86 may be useful for prognostic assessment in patients with NSCLC.
Conclusions
In summary, we provide the first clinical evidence that C9orf86 can be detected in a subset of NSCLC and that its expression is related to lymph node metastasis and clinical stage. We demonstrated that C9orf86 expression is a prognostic indicator of poor survival among patients with NSCLC; thus, C9orf86 may be useful as a biomarker to determine the prognosis of NSCLC and is a potential therapeutic target for NSCLC treatment.
Acknowledgements
Funding: This study was supported by grants from the National Natural Science Foundation of China (No. 8120184).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The experiment was approved by the Institutional Ethics Committee of Guangzhou Medical University (ID: 2015, No.21).
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Zou X. Epidemiology of lung cancer in China. Chin J Cancer Preve Treat 2007;14:881-3.
- Paesmans M, Sculier JP, Libert P, et al. Prognostic factors for survival in advanced non-small-cell lung cancer: univariate and multivariate analyses including recursive partitioning and amalgamation algorithms in 1,052 patients. The European Lung Cancer Working Party. J Clin Oncol 1995;13:1221-30. [PubMed]
- Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med 2004;350:351-60. [Crossref] [PubMed]
- Kelly EE, Horgan CP, Goud B, et al. The Rab family of proteins: 25 years on. Biochem Soc Trans 2012;40:1337-47. [Crossref] [PubMed]
- Hernández-Alcoceba R, del Peso L, Lacal JC. The Ras family of GTPases in cancer cell invasion. Cell Mol Life Sci 2000;57:65-76. [Crossref] [PubMed]
- Saez R, Chan AM, Miki T, et al. Oncogenic activation of human R-ras by point mutations analogous to those of prototype H-ras oncogenes. Oncogene 1994;9:2977-82. [PubMed]
- Graham SM, Cox AD, Drivas G, et al. Aberrant function of the Ras-related protein TC21/R-Ras2 triggers malignant transformation. Mol Cell Biol 1994;14:4108-15. [Crossref] [PubMed]
- Luo H, Hao X, Ge C, et al. TC21 promotes cell motility and metastasis by regulating the expression of E-cadherin and N-cadherin in hepatocellular carcinoma. Int J Oncol 2010;37:853-9. [PubMed]
- Yu L, Hui-chen F, Chen Y, et al. Differential expression of RAB5A in human lung adenocarcinoma cells with different metastasis potential. Clin Exp Metastasis 1999;17:213-9. [Crossref] [PubMed]
- Liu SS, Chen XM, Zheng HX, et al. Knockdown of Rab5a expression decreases cancer cell motility and invasion through integrin-mediated signaling pathway. J Biomed Sci 2011;18:58. [Crossref] [PubMed]
- Schnelzer A, Prechtel D, Knaus U, et al. Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 2000;19:3013-20. [Crossref] [PubMed]
- Cheng KW, Lahad JP, Kuo WL, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med 2004;10:1251-6. [Crossref] [PubMed]
- Fan Y, Xin XY, Chen BL, et al. Knockdown of RAB25 expression by RNAi inhibits growth of human epithelial ovarian cancer cells in vitro and in vivo. Pathology 2006;38:561-7. [Crossref] [PubMed]
- Li Q, Wang L, Zeng L, et al. Evaluation of the novel gene Rabl3 in the regulation of proliferation and motility in human cancer cells. Oncol Rep 2010;24:433-40. [PubMed]
- Montalbano J, Lui K, Sheikh MS, et al. Identification and characterization of RBEL1 subfamily of GTPases in the Ras superfamily involved in cell growth regulation. J Biol Chem 2009;284:18129-42. [Crossref] [PubMed]
- Montalbano J, Jin W, Sheikh MS, et al. RBEL1 is a novel gene that encodes a nucleocytoplasmic Ras superfamily GTP-binding protein and is overexpressed in breast cancer. J Biol Chem 2007;282:37640-9. [Crossref] [PubMed]
- Li YY, Fu S, Wang XP, et al. Down-regulation of c9orf86 in human breast cancer cells inhibits cell proliferation, invasion and tumor growth and correlates with survival of breast cancer patients. PLoS One 2013;8:e71764. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw 2013;11:645-53. [PubMed]
- Finn RS, Press MF, Dering J, et al. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol 2009;27:3908-15. [Crossref] [PubMed]
- Mitsuuchi Y, Testa JR. Cytogenetics and molecular genetics of lung cancer. Am J Med Genet 2002;115:183-8. [Crossref] [PubMed]
- Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet 2002;3:101-28. [Crossref] [PubMed]
- Jaffee EM, Hruban RH, Canto M, et al. Focus on pancreas cancer. Cancer Cell 2002;2:25-8. [Crossref] [PubMed]
- Mazieres J, Antonia T, Daste G, et al. Loss of RhoB expression in human lung cancer progression. Clin Cancer Res 2004;10:2742-50. [Crossref] [PubMed]
- Ma D, Bai X, Zou H, et al. Rheb GTPase controls apoptosis by regulating interaction of FKBP38 with Bcl-2 and Bcl-XL. J Biol Chem 2010;285:8621-7. [Crossref] [PubMed]
- Wang JS, Wang FB, Zhang QG, et al. Enhanced expression of Rab27A gene by breast cancer cells promoting invasiveness and the metastasis potential by secretion of insulin-like growth factor-II. Mol Cancer Res 2008;6:372-82. [Crossref] [PubMed]
- Mitra S, Cheng KW, Mills GB. Rab25 in cancer: a brief update. Biochem Soc Trans 2012;40:1404-8. [Crossref] [PubMed]


