Mutations and expression of the NFE2L2/KEAP1/CUL3 pathway in Chinese patients with lung squamous cell carcinoma
Introduction
As the leading cause of cancer-related deaths, lung cancer causes an estimated 1.6 million deaths worldwide and 0.5 million deaths in China every year (1,2). The discovery in recent years of mutations or fusions involving the epidermal growth factor receptor (EGFR) kinase and anaplastic lymphoma kinase (ALK), has greatly improved the treatment of patients with lung adenocarcinoma, one of the two most common subtypes of lung cancer (3-6). However, these targeted agents, which were developed for lung adenocarcinoma are largely ineffective against lung squamous cell carcinoma (SqCC), the other most common subtype of lung cancer (7,8).
In recent years, several studies have reported an abnormally high alteration rate in the nuclear factor erythroid 2-like 2 (NFE2L2)/kelch-like ECH-associated protein 1 (KEAP1)/cullin 3 (CUL3) pathway in many types of cancers, including lung SqCC (9-11). Regulating the response to oxidative stress, NFE2L2 is a master transcriptional activator of genes which contain antioxidant response elements (AREs) in their promoters and in response to oxidative stress. In normal cells, KEAP1 binds to NFE2L2 and NFE2L2 is polyubiquitylated by the CUL3-based E3 ligase complex, resulting in rapid NFE2L2 degradation by proteasomes at the baseline, while under oxidative stress KEAP1 is modified, which leads to the inhibition of NFE2L2 ubiquitylation (12,13). But in tumors, the frequently occurring mutations of NFE2L2 or KEAP1 will cause disruption of their normal combination (14). As a result, NFE2L2 cannot pass through ubiquitination and degradation; finally the intracellular accumulation of NFE2L2 will bring about the activation of its downstream genes, increase resistance to oxidative stress, and promote tumor growth (14).
At present, the status of the NFE2L2/KEAP1/CUL3 pathway in Chinese patients with lung SqCC has not been determined. Thus, we investigated the mutations and expression of this pathway in tumors and normal tissues obtained from Chinese patients with lung SqCC. We identified several unreported mutations, and confirmed that NFE2L2 and KEAP1 were both up-regulated in lung SqCC compared with adjacent normal samples, while the expression of CUL3 needs further research. Our results will promote an understanding of this pathway and that this knowledge will benefit the treatment of Chinese patients with lung SqCC.
Methods
Tissue samples
As previously reported (15), samples were obtained from patients with lung SqCC who underwent surgical resection between July and December, 2012 at Zhongshan Hospital, Fudan University, Shanghai, China. All samples were quickly frozen in liquid nitrogen after extirpation and then stored at –80 °C. Finally, a total of 100 pairs of normal tissue samples and lung SqCC tissues were obtained, and then used in sequencing and RT-qPCR. Paraffin-embedded specimens obtained from 50 other patients were used in immunohistochemical staining.
cDNA preparation
As previously reported (15), total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) and integrity was evaluated using agarose gel electrophoresis. DNA contamination was eliminated using gDNA Eraser (TaKaRa, Tokyo, Japan), and then cDNA synthesis was performed using the PrimeScriptTM RT Master Mix (Perfect Real Time; TaKaRa).
cDNA sequencing
First, PCR was carried out using AmpliTaq Gold 360 Master Mix (ABI, Foster City, CA, USA) with the following procedure: 1 cycle at 95 °C for 10 mins; 40 cycles at 94 °C for 30 secs, at 60 °C for 30 secs, at 72 °C for 40 secs; 1 cycle at 72 °C for 10 mins. The tagged primers used in this step were synthetized by Life Technologies (New York, NY, USA) and efficiencies were determined by gel electrophoresis. The PCR products were then purified using ExoSAP-IT PCR Product Cleanup (Affymetrix, Santa Clara, CA, USA), prepared using BigDye Terminator V3.1 Cycle Sequencing Kit (ABI), and then sequenced using a Prism 3700 DNA Analyzer (ABI) in duplicate according to the manufacturer’s guidelines. Sequencing results were compared with corresponding entries in the National Centre for Biotechnology Information (NCBI) Nucleotide Database (http://www.ncbi.nlm.nih.gov/nuccore/, NFE2L2: NM_006164.4; KEAP1: NM_203500.1; CUL3: NM_003590.4). All of the mutations detected were further confirmed by reduplicated experiments. Single nucleotide polymorphism (SNP) information was obtained from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/snp/).
TaqMan qPCR
qPCR reactions were carried out in triplicate using TaqMan Gene Expression Master Mix (Life Technologies) with the following PCR procedure: 1 cycle at 95 °C for 10 mins; 40 cycles at 95 °C for 15 secs and at 60 °C for 1 min. The primers and TaqMan probes of NFE2L2 (Hs00975961_g1), KEAP1 (Hs00202227_m1), CUL3 (Hs00180183_m1), and β-actin (Hs01060665_g1) were provided by Life Technologies; β-actin was applied as an endogenous control to eliminate experimental errors. The 2−ΔΔCT method was used to calculate the expression of these genes.
Immunohistochemical staining
As previously reported (16), immunohistochemical staining was performed with EnVisionTM HRP-polymer anti-mouse/rabbit IHC Kit (KeyGEN BioTECH, Nanjing, Jiangsu, China). Briefly, the antibodies specific for NFE2L2 (1:200 dilution, Abcam, Cambridge, UK), KEAP1 (1:200 dilution, Abcam) and CUL3 (1:200 dilution, Abcam) were used to detect these three proteins. The levels of expression were assessed semi-quantitatively as the percentage of marked target cells and the staining intensity as recommended previously (17). Finally, we separated the specimens according to expression in four groups (negative, weak, moderate, and strong).
Statistical analysis
The data were analyzed with IBM SPSS for Windows (version 20, IBM, Armonk, NY, USA). T-test, Mann-Whitney U test, and Wilcoxon signed rank test were used to evaluate the differences in the levels of expression. A P value <0.05 was considered statistically significant.
Results
NFE2L2, KEAP1, and CUL3 mutations
As Table S1 shows, 47 mutations were detected in 36 patients (36%). Twenty-three NFE2L2 mutations were detected in 17 patients (17%), including 21 missense mutations. Thirteen missense KEAP1 mutations, including one deletion, were detected in of 12 patients (12%), while four synonymous and seven missense CUL3 mutations, including two deletions and one stop-gain mutation, were detected in CUL3 of 10 patients (10%).
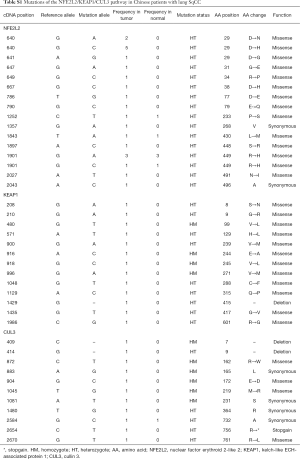
Full table
Most of these mutations were only detected in the SqCC samples but not the paired normal samples, with the exception of six NFEL2 and one CUL3 mutations. Thirty-seven mutations (78.7%) were heterozygous, thus both mutated and normal proteins were expressed in cancer cells. We showed that the 29th amino acid (AA) of NFE2L2 was mutated in eight patients (8%) and the 449th AA of NFE2L2 in four patients (4%). The mutation rates in these two sites were considerable, and the results of the following AA and function changes are possibly worth further study.
One hundred and forty-three SNPs were detected in 59 patients (59%), all of these SNPs occurred in KEAP1 and CUL3, but not NFE2L2 (Table 1). All of these SNPs were detected in the SqCC samples and the paired normal samples simultaneously. Only the rs3738952 SNP detected in 31 patients (31%, 7 homozygotes and 24 heterozygotes) resulted in an AA change of CUL3; it is not clear how the functional change occurred.
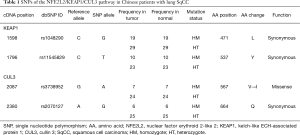
Full table
NFE2L2, KEAP1, and CUL3 expression
We first detected NFE2L2, KEAP1, and CUL3 expression by RT-qPCR using TaqMan probes in 100 pairs of lung SqCC samples and precarcinomatous normal tissues. As shown in Figure 1, gene expressions were all significantly upregulated in lung SqCC samples compared with corresponding normal samples (NFE2L2, 2.76-fold, P<0.001; KEAP1 2.73-fold, P<0.001; CUL3, 1.25-fold, P=0.010; paired t-test).

To further fix the expressions of the three genes, we used immunohistochemistry staining to measure gene expressions in 50 lung SqCC specimens. As Table 2 shows, NFE2L2 expression was significantly higher in lung SqCC (P<0.001, Wilcoxon signed-rank test), which was consistent with the RT-qPCR results, while KEAP1 expression was also elevated in most lung SqCC samples, but not significantly (P=0.056, Wilcoxon signed rank test). There was no significant difference in CUL3 expression in lung SqCC or normal samples (P=0.996, Wilcoxon signed-rank test).
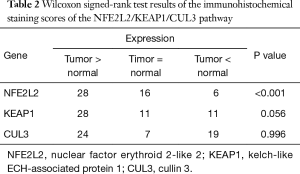
Full table
Based on the RT-qPCR and immunohistochemistry results, we confirmed that NFE2L2 was more highly expressed in lung SqCC than in normal lung tissues. Even though the statistical analysis of KEAP1 immunohistochemistry staining was not significant, we still believed that KEAP1 was also up-regulated in lung SqCC in consideration of its P value slightly higher than 0.05 and the RT-qPCR results. The RT-qPCR results showed that the gap of the CUL3 expression in lung SqCC and normal tissue was marginal, and the difference was not detected in immunohistochemistry staining, thus we considered that the expression of CUL3 in lung SqCC needed further evaluation.
Discussion
In this research, we reported a number of mutations and SNPs of the NFE2L2/KEAP1/CUL3 pathway in more than half of Chinese patients with lung SqCC, many of which will lead to an alteration in the normal AA sequence. However, the subsequent function change caused by each mutation and SNP remains elusive and needs to be further studied. In addition, combining RT-qPCR and immunohistochemistry staining, we confirmed that the expression of NFE2L2 and KEAP1 were elevated while the expression of CUL3 was not significantly changed in Chinese patients with lung SqCC.
A number of studies have reported the abnormal incidence of mutations in the NFE2L2/KEAP1/CUL3 pathway and high expression of NFE2L2 in various cancers, in agreement with our results (9,18-21). Nevertheless, there are some controversial views about the expression of KEAP1 in tumorigenesis. Solis et al. (22) reported that the expression of KEAP1 is decreased in 56% of non-small cell lung cancers. And Chien et al. (23) reported that KEAP1 expression is decreased in specimens from NSCLC patients with lymph node metastases compared with patients without metastasis. But Huang et al. (24,25) concluded that KEAP1 is up-regulated in patients with oral squamous cell and salivary adenoid cystic carcinomas, and associated with carcinogenesis and progression. Few studies have focused on the expression of CUL3, only Haagenson et al. (26) has reported that CUL3 is increasingly expressed during progression from early breast cancer to invasive carcinoma.
Several tumor-associated proteins have been shown to inhibit the degradation of NFE2L2, such as p62, Kirsten rat sarcoma viral oncogene homolog (KRAS), sirtuin 5 (SIRT5), and transforming growth factor (TGF) β, then promote cell proliferation and chemotherapeutic resistance (27-30). The NFE2L2/KEAP1/CUL3 pathway has been reported to interact with many other important signaling pathways, including phosphatidylinositol-4,5-bisphosphate 3-kinase/v-akt murine thymoma viral oncogene homolog 1 (PI3K/Akt), NOTCH, nuclear factor (NF)-κB, and extracellular regulated mitogen-activated protein kinase (ERK), thus comprising a complex regulation network within cancer cells (31-36).
Conclusions
In sum, our results suggested that the NFE2L2/KEAP1/CUL3 pathway was frequently mutated and abnormally expressed in Chinese lung SqCC. We hope our results will help other researchers to find new therapies targeted at its abnormal mutations or expressions. Due to the important function of the NFE2L2/KEAP1/CUL3 pathway in cancer cells, it can be concluded that this pathway will play an important role in tumor therapy.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China (Grant Nos. 81401875, 81472225) (http://www.nsfc.gov.cn/) and the Natural Science Foundation of Shanghai, China (Grant No. 14ZR1406000) (http://www.stcsm.gov.cn/). And we would like to thank the language editing service, ICE editing Co for editing this manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University {No. 2011-219[2]} and written informed consent was obtained from all patients, who participated in this research when they were hospitalized.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Chen W, Zheng R, Zhang S, et al. Report of cancer incidence and mortality in China, 2010. Ann Transl Med 2014;2:61. [PubMed]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [Crossref] [PubMed]
- Yan M, Parker BA, Schwab R, et al. HER2 aberrations in cancer: implications for therapy. Cancer Treat Rev 2014;40:770-80. [Crossref] [PubMed]
- Sui X, Kong N, Zhu M, et al. Cotargeting EGFR and autophagy signaling: A novel therapeutic strategy for non-small-cell lung cancer. Mol Clin Oncol 2014;2:8-12. [PubMed]
- Iwama E, Okamoto I, Harada T, et al. Development of anaplastic lymphoma kinase (ALK) inhibitors and molecular diagnosis in ALK rearrangement-positive lung cancer. Onco Targets Ther 2014;7:375-85. [PubMed]
- Cooper WA, Lam DC, O'Toole SA, et al. Molecular biology of lung cancer. J Thorac Dis 2013;5 Suppl 5:S479-90. [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [Crossref] [PubMed]
- Kobayashi A, Kang MI, Watai Y, et al. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 2006;26:221-9. [Crossref] [PubMed]
- Kobayashi A, Kang MI, Okawa H, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 2004;24:7130-9. [Crossref] [PubMed]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 2009;34:176-88. [Crossref] [PubMed]
- Zhan C, Zhang Y, Ma J, et al. Identification of reference genes for qRT-PCR in human lung squamous-cell carcinoma by RNA-Seq. Acta Biochim Biophys Sin (Shanghai) 2014;46:330-7. [Crossref] [PubMed]
- Zhan C, Yan L, Wang L, et al. Identification of immunohistochemical markers for distinguishing lung adenocarcinoma from squamous cell carcinoma. J Thorac Dis 2015;7:1398-405. [PubMed]
- Zhan C, Shi Y, Lu C, et al. Pyruvate kinase M2 is highly correlated with the differentiation and the prognosis of esophageal squamous cell cancer. Dis Esophagus 2013;26:746-53. [PubMed]
- Sasaki H, Suzuki A, Shitara M, et al. Genotype analysis of the NRF2 gene mutation in lung cancer. Int J Mol Med 2013;31:1135-8. [PubMed]
- Hartikainen JM, Tengström M, Kosma VM, et al. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res 2012;72:5537-46. [Crossref] [PubMed]
- Onodera Y, Motohashi H, Takagi K, et al. NRF2 immunolocalization in human breast cancer patients as a prognostic factor. Endocr Relat Cancer 2014;21:241-52. [Crossref] [PubMed]
- Ji L, Wei Y, Jiang T, et al. Correlation of Nrf2, NQO1, MRP1, cmyc and p53 in colorectal cancer and their relationships to clinicopathologic features and survival. Int J Clin Exp Pathol 2014;7:1124-31. [PubMed]
- Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 2010;16:3743-53. [Crossref] [PubMed]
- Chien MH, Lee WJ, Hsieh FK, et al. Keap1-Nrf2 Interaction Suppresses Cell Motility in Lung Adenocarcinomas by Targeting the S100P Protein. Clin Cancer Res 2015;21:4719-32. [Crossref] [PubMed]
- Huang CF, Deng WW, Zhang L, et al. Expression of LC3, LAMP2, KEAP1 and NRF2 in Salivary Adenoid Cystic Carcinoma. Pathol Oncol Res 2016;22:109-14. [Crossref] [PubMed]
- Huang CF, Zhang L, Ma SR, et al. Clinical significance of Keap1 and Nrf2 in oral squamous cell carcinoma. PLoS One 2013;8:e83479. [Crossref] [PubMed]
- Haagenson KK, Tait L, Wang J, et al. Cullin-3 protein expression levels correlate with breast cancer progression. Cancer Biol Ther 2012;13:1042-6. [Crossref] [PubMed]
- Tao S, Wang S, Moghaddam SJ, et al. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res 2014;74:7430-41. [Crossref] [PubMed]
- Oshimori N, Oristian D, Fuchs E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 2015;160:963-76. [Crossref] [PubMed]
- Sun X, Ou Z, Chen R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016;63:173-84. [Crossref] [PubMed]
- Lu W, Zuo Y, Feng Y, et al. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol 2014;35:10699-705. [Crossref] [PubMed]
- Wakabayashi N, Chartoumpekis DV, Kensler TW. Crosstalk between Nrf2 and Notch signaling. Free Radic Biol Med 2015;88:158-67. [Crossref] [PubMed]
- Wakabayashi N, Skoko JJ, Chartoumpekis DV, et al. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol 2014;34:653-63. [Crossref] [PubMed]
- Reddy NM, Potteti HR, Vegiraju S, et al. PI3K-AKT Signaling via Nrf2 Protects against Hyperoxia-Induced Acute Lung Injury, but Promotes Inflammation Post-Injury Independent of Nrf2 in Mice. PLoS One 2015;10:e0129676. [Crossref] [PubMed]
- Chuang JI, Huang JY, Tsai SJ, et al. FGF9-induced changes in cellular redox status and HO-1 upregulation are FGFR-dependent and proceed through both ERK and AKT to induce CREB and Nrf2 activation. Free Radic Biol Med 2015;89:274-86. [Crossref] [PubMed]
- Huang CS, Lin AH, Yang TC, et al. Shikonin inhibits oxidized LDL-induced monocyte adhesion by suppressing NFκB activation via up-regulation of PI3K/Akt/Nrf2-dependent antioxidation in EA.hy926 endothelial cells. Biochem Pharmacol 2015;93:352-61. [Crossref] [PubMed]
- Harada S, Nakagawa T, Yokoe S, et al. Autophagy Deficiency Diminishes Indomethacin-Induced Intestinal Epithelial Cell Damage through Activation of the ERK/Nrf2/HO-1 Pathway. J Pharmacol Exp Ther 2015;355:353-61. [Crossref] [PubMed]


