Long noncoding RNAs are novel potential prognostic biomarkers for esophageal squamous cell carcinoma: an overview
Introduction
Esophageal cancer is the eighth most common malignant tumors worldwide and the sixth most common cause of death from cancer with geographic differences in incidence rate (1). Histologically, esophageal cancer mainly consists of squamous cell carcinoma and adenocarcinoma. In highest-incidence areas, such as in China, more than 90% of the esophageal cancers are esophageal squamous cell carcinoma (ESCC) (2). Despite the advancement in treating ESCC, the overall survival still remains poor, with a five-year survival rate of 15–34% (1,3,4). To date, the factors that influence the development and prognosis of ESCC are still not clearly known (5). Therefore, identification of the biomarkers which could predict the prognosis of ESCC remains meaningful and urgently necessary, which can be not only helpful with clinical monitoring but also suggestive of therapeutic decision-making (6).
Recently, long noncoding RNAs (lncRNAs), which are commonly defined as non-protein-coding RNA molecule longer than 200 nucleotides, have been intensively researched all around the world. LncRNAs are a new group of noncoding RNAs locating within nuclear or cytosolic fractions (7), which were found to be important regulators of tissue physiology and diseases processes, especially in cancer (8). To date, more than 5,000 human lncRNAs have been identified via high-throughput techniques (9). There are several kinds of lncRNAs mainly consisting of intergenic RNAs, intronic RNAs, sense lncRNAs, antisense lncRNAs and others (7,10,11), many of which are transcribed by RNA polymerase II and polyadenylated. Both lncRNAs and small noncoding RNAs make up the scope of noncoding RNAs (10,12), and the cut-off length for these two groups is 200 nucleotides. Small noncoding RNAs such as miRNAs, siRNAs, and tRNAs have already been well and widely researched (13), while lncRNAs as one of the most common RNA species are poorly understood (14). However, with the development of whole genome and transcriptome sequencing technologies, recent studies have found that lncRNAs played an important role not only between DNA and protein but also in cellular functions (12). Moreover, lncRNAs were recently found to be closely related to cancer including ESCC. Therefore, in this review, we tried to summarize the relation between lncRNAs and ESCC as well as the biomarker value of lncRNAs in ESCC.
LncRNAs and cancer: an emerging noncoding RNA closely related to cancer
Cancer has been known to have the ability to sustain proliferate signaling and evade growth suppressors, and consequently resist cell death (15). It is well known that tumorigenesis is related with various deregulations of different biomolecules and signal pathways. However, the detailed mechanism of initiation and progression of cancer still needs to be uncovered and clearly understood. Recently, aberrant expression of the lncRNAs was found to be closely related to human diseases, especially cancer. Therefore, more and more studies have focused on lncRNAs, which were found to lead to cancer development and progression through multiple mechanisms (16). Recent evidence has begun to accumulate describing the mechanisms of the lncRNAs related to cancer, and these mechanisms mainly involved in epigenetics, transcription regulation, and post-transcription processing (9,12,16,17). Generally, some lncRNAs (oncogenic lncRNAs) play an oncogenic role in cancer, while others (tumor suppressor lncRNAs) have oncosuppressive functions (9,15). With the development of various techniques including expression microarrays, tiling arrays, next generation sequencing, and methylation analysis (18), recent studies have revealed many oncogenic lncRNAs, for example, HOX antisense intergenic RNA (HOTAIR), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), steroid receptor RNA activator (SRA), and antisense non-coding RNA in the INK4 locus (ANRIL), as well as some oncosuppressive lncRNAs such as maternally expressed gene 3 (MEG3), growth arrest-specific 5 (GAS5), telomeric repeat-containing RNA (TERRA), and CCND1/Cyclin D1 (9,15,17). The detailed mechanisms of how those lncRNAs were related to cancer have been intensively discussed [see (17-21) for detailed reviews]. Dysregulation of lncRNAs played critical roles in tumorigenesis and tumor progression, and lncRNAs may become potential biomarkers in various types of cancer including ESCC (22). Accumulating evidence showed that dysregulation of lncRNAs could promote ESCC cell proliferation and metastasis, indicating that abnormal expression of lncRNAs is significantly related to the poor prognosis of patients with ESCC (6,23-26). Therefore, lncRNAs may be a group of prognostic markers for ESCC. Herein, our current review specifically focused on the relationship of lncRNAs and ESCC.
LncRNAs and ESCC: the roles of lncRNAs in ESCC and their prognostic value
Even though there are a lot of researches demonstrating the mechanisms of the development and progression of ESCC (27), there are still much more to uncover and understand, and the prognosis of ESCC remains dismal. Therefore, it urgently requires the identification of biomarkers which may explain the mechanisms of tumorigenesis and predict the prognosis for clinical monitoring as well as therapeutic strategies of ESCC. With the development of biomolecular and genetic techniques, many researches have been carried out to explore the relationship and potential mechanisms between various lncRNAs and ESCC, as well as the prognostic value of lncRNAs in ESCC (5,6,23-26,28-33). These major lncRNAs with aberrant expression in ESCC were summarized in Table 1.
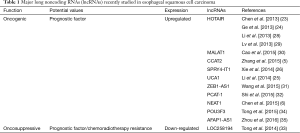
Full table
HOTAIR, as a predominantly focused oncogenic lncRNA (36), was initially found to be involved with primary breast tumor and its metastasis (23,37). It is widely known that overexpression of HOTAIR induced genome-wide re-targeting of polycomb repressive complex 2 (PRC2), which led to an altered methylation of histone H3 lysine 27 (H3K27) and gene expression (especially metastasis-suppressing genes), and thus the aberrant expression of HOTAIR promoted tumor invasiveness and metastasis (23,24,28,37). Recently, accumulating evidence showed that HOTAIR was related closely to the development and progression of ESCC (38). Studies have found that HOTAIR was notably highly expressed in ESCC tumor tissues and the overexpression of HOTAIR was significantly related to poor prognosis of the patients (23,24,28,29). Studies have showed that elevated HOTAIR expression was correlated with increased occurrence of lymph node metastasis and shorter overall survival (23,24,28,29). HOTAIR was believed to act its role in promoting the invasiveness and progression of ESCC in a manner dependent on PRC2 (38), which was also showed to inhibit WIF-1 expression and activate Wnt pathway (24). In vitro, knockdown or suppression of the HOTAIR reduced the invasiveness and metastasis of ESCC cells (23,28). The detailed mechanisms, however, still need further investigations to elucidate. Based on above evidence, it is believed that HOTAIR remained to be an independent prognostic factor of ESCC as well as potential biomarker for the existence of lymph node metastasis in ESCC. A recent meta-analysis also found that elevated level of HOTAIR was a powerful prognostic biomarker for patients with ESCC and may serve as a potential therapeutic target for ESCC (39), which may lead the direction of the researches on novel therapeutic strategy for ESCC.
MALAT1, as one of the most studied oncogenic lncRNAs, was originally showed to be overexpressed in non-small cell lung cancer patients with lymph node metastasis (40). However, apart from lung cancer, MALAT1 was found to be closely related to various cancers (bladder cancer, gallbladder carcinoma, prostate cancer), and significantly correlated with relapse and progression of these cancers (15). Recent evidence showed that MALAT1 promoted tumor growth by regulating cell cycle and epithelial-to-mesenchymal transition as well as angiogenesis (10), which may lead to uncontrollable tumor growth. MALAT1 was also found to be overexpressed in ESCC cells (30,41), and its aberrant expression was positively correlated with clinical stage and lymph node metastasis (41), as well as poor prognosis (30) of ESCC patients. Hu et al. (41) have showed that up-regulation of MALAT1 may promote ESCC growth by dephosphorylation of the ATM-CHK2 pathway, which may lead to the loss of cell cycle arrest and ultimately proliferation and metastasis of ESCC cells. Recent studies have showed that knockdown or silencing of MALAT1 can lead to G2/M phase arrest and an increased apoptosis ratio (41,42), and thus inhibit proliferation, migration, and invasion of ESCC cells (43). Moreover, Wang et al. (44) found that knockdown of MALAT1 decreased tumor formation and improved survival in animal experiments, suggesting that inhibition of MALAT1 may be a potential target for treating ESCC. However, how the amplification of MALAT1 in ESCC cells occurs still remains unclear. Therefore, further studies are warranted to explore the prognostic biomarker role of MALAT1 in ESCC as well as its potentiality of therapeutic target for treating ESCC.
Colon cancer-associated transcript 2 (CCAT2) is a novel lncRNA, which was found to be related with colon cancer (45), breast cancer (46), lung cancer (47), gastric cancer (48), as well as ESCC (5). In ESCC, the level of CCAT2 expression was positively related with TNM stage and lymph node metastasis, and high expression of CCAT2 correlated with poor survival of ESCC patients (5). Similarly, Wang et al. found that CCAT2 was significantly overexpressed in ESCC tumor tissue and that CCAT2 might have the potential as a diagnostic biomarker (49). Based on above evidence, CCAT2 could also serve as a prognostic as well as diagnostic biomarker and a therapeutic target of ESCC, and it is worthy of further study.
With more and more researches exploring the relationship between lncRNAs and ESCC accomplished, many other lncRNAs were continuously found out to be significantly related to ESCC. Other overexpressed lncRNAs included lncRNA SPRY4 intronic transcript 1 (SPRY4-IT1) (26,50), urothelial carcinoma associated 1 (UCA1) (25), lncRNA ZEB1 anti-sense1 (ZEB1-AS1) (31), prostate cancer-associated ncRNA transcript 1 (PCAT-1) (32), nuclear paraspeckle assembly transcript 1 (NEAT1) (6), and lncRNA AFAP1-AS1 (35). The upregulation of above lncRNAs was significantly related to poor prognosis of patients with ESCC, and those lncRNAs could be potential therapeutic target of ESCC (summarized in Table 1).
LncRNAs have not only oncogenic functions but also oncosuppressive roles. LncRNA LOC258194, also called LSAMP antisense RNA 3, was showed to be within a tumor suppressor unit in osteosarcoma and to suppress tumor cell growth (51), and was found to be a p53-regulated oncosuppressive lncRNA (52). Recently, Tong et al. (33) found that the expression of lncRNA LOC285194 was significantly down-regulated in ESCC tumor tissue as compared with normal esophageal tissue, and low expression of lncRNA LOC285194 was significantly related to advanced TNM stage and metastasis of ESCC as well as poor prognosis of patients with ESCC. Moreover, low expression of lncRNA LOC285194 was also associated with chemoradiotherapy resistance. Therefore, decreased expression of lncRNA LOC285194 may serve as a biomarker of prognosis as well as selection criteria for patients who could benefit from chemoradiotherapy (33). Recently, Wang et al. (53) have introduced another oncosuppressive lncRNA in ESCC, lncRNA-Low Expression in Tumor (lncRNA-LET). They found that the expression level of lncRNA-LET was decreased in ESCC tissue and overexpression of lncRNA-LET could inhibit the migration and invasion of ESCC cells in vitro, suggesting that lncRNA-LET may serve as a tumor suppressor in ESCC (53). However, to date, oncosuppressive lncRNAs were hardly explored and figured out in ESCC. Further studies are needed to figure out the oncosuppressive functions of lncRNAs in ESCC, which may help with better understanding of development and progression of ESCC.
Future prospective
As accumulating evidence has showed that lncRNAs played an important role in the tumorigenesis and progression of ESCC and that lncRNAs may serve as a prognostic biomarker as well as a therapeutic target for ESCC, more researches are warranted to unveil the detailed relationship between lncRNAs and ESCC. However, as a novel field for further studies, there are some future prospective needed to be addressed. First, although more and more lncRNAs were figured out to be related to ESCC, the underlying mechanisms of how those lncRNAs correlated to ESCC are still unknown, and therefore, the therapeutic value of lncRNAs is still theoretical. As a result, the bench research-testified therapeutic benefits of lncRNAs in ESCC are still at its very beginning, and how to transfer those bench research benefits into clinical benefits is another important aspect for future researches. Second, most of the lncRNAs mentioned above were evaluated in ESCC tumor tissue, and those lncRNAs showed the prognostic value for patients with ESCC. However, similar to miRNAs, lncRNAs could also serve as a diagnostic biomarker for ESCC (22). So far, only Wang et al. (49) and Tong et al. (34) have explored the diagnostic value of lncRNAs in ESCC. Tong et al. (34) found that lncRNA POU3F3 in plasma could serve as a potential noninvasive biomarker for diagnosis of ESCC, which may help with early tumor screening. Therefore, the noninvasive diagnostic value of lncRNAs in ESCC is still needed for further researches to elucidate. Third, as there are more and more new lncRNAs being found out to significantly correlate with ESCC, how to improve the prognostic and diagnostic power of lncRNAs for ESCC still remains to be solved. In order to robustly predict the survival of patients with ESCC, Li et al. (54) have firstly found out that a three-lncRNA signature (including the lncRNAs ENST00000435885.1, XLOC_013014 and ENST00000547963.1) could serve as a new biomarker for the prognosis of patients with ESCC. Similar to miRNAs, a three-lncRNA signature enabled more accurate prediction of survival for those patients. Similarly, Pan et al. (55) used “lncRNA-mRNA gene pair” (lncRNA FOXCUT and mRNA FOXC1 pair) to predict the prognosis of patients with ESCC, and they found that this novel lncRNA-mRNA pair could represent a potential prognostic biomarker and therapeutic target for ESCC patients. Therefore, a combination of lncRNAs or lncRNAs and other genes could be used as a more accurate prognostic biomarker for patients with ESCC. Future studies are needed, however, to explore the prognostic and diagnostic value of such combination in ESCC before their biomarker functions can be applied in clinical practice. Finally, most of those researches concerned predominantly the roles of lncRNAs in ESCC, while only a small number of studies focused on esophageal adenocarcinoma (56) which is the main pathologic type of esophageal cancer in western countries. As lncRNAs have showed more and more promising results in ESCC, more efforts are needed to explore the role of lncRNAs in esophageal adenocarcinoma.
Conclusions
Patients with ESCC still have a poor prognosis, and identification of prognostic and diagnostic biomarkers for ESCC is still badly needed. With the development of genome and transcriptome sequencing technologies, more and more dysregulated oncogenic and oncosuppressive lncRNAs were found out to be closely related to various cancers. There are also a myriad of published studies about aberrant expression of lncRNAs in ESCC. Even though current knowledge has revealed the possible mechanisms between lncRNAs and cancers (including epigenetics, transcription regulation, and posttranscription processing), the detailed mechanisms of lncRNAs in the tumorigenesis and progression of ESCC are still unclear. In our current review, we summarized the prognostic value of lncRNAs in ESCC, and pointed out that lncRNAs may serve as novel prognostic and diagnostic biomarkers as well as therapeutic targets for ESCC. Our study highlighted the promising biomarker functions of lncRNAs in ESCC and pointed out some perspectives needed to be addressed for future researches. Even though there is still a long way from bench research-testified benefits to clinical benefits, with more and more researches digging into the aberrant expression of lncRNAs in ESCC, we believe that lncRNAs will exert more important roles in the treatment of ESCC in the near future.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400-12. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Sjoquist KM, Burmeister BH, Smithers BM, et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol 2011;12:681-92. [Crossref] [PubMed]
- Fakhrian K, Ordu AD, Lordick F, et al. Long-term outcomes of trimodality treatment for squamous cell carcinoma of the esophagus with cisplatin and/or 5-FU. Strahlentherapie und Onkologie 2014;190:1133-40. [Crossref] [PubMed]
- Zhang X, Xu Y, He C, et al. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol 2015;111:834-9. [Crossref] [PubMed]
- Chen X, Kong J, Ma Z, et al. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res 2015;5:2808-15. [Crossref] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. [Crossref] [PubMed]
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199-208. [Crossref] [PubMed]
- Meseure D, Drak Alsibai K, Nicolas A, et al. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. Biomed Res Int 2015;2015:320214.
- Silva A, Bullock M, Calin G. The Clinical Relevance of Long Non-Coding RNAs in Cancer. Cancers (Basel) 2015;7:2169-82. [Crossref] [PubMed]
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet 2015;31:239-51. [Crossref] [PubMed]
- Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013;339:159-66. [Crossref] [PubMed]
- Donzelli S, Cioce M, Muti P, et al. MicroRNAs: Non-coding fine tuners of receptor tyrosine kinase signalling in cancer. Semin Cell Dev Biol 2016;50:133-42. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Hajjari M, Khoshnevisan A, Shin YK. Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumour Biol 2014;35:10645-63. [Crossref] [PubMed]
- Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol (Dordr) 2015;38:17-28. [Crossref] [PubMed]
- Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci 2013;14:4655-69. [Crossref] [PubMed]
- Mitra SA, Mitra AP, Triche TJ. A central role for long non-coding RNA in cancer. Front Genet 2012;3:17. [Crossref] [PubMed]
- Cheetham SW, Gruhl F, Mattick JS, et al. Long noncoding RNAs and the genetics of cancer. Br J Cancer 2013;108:2419-25. [Crossref] [PubMed]
- Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 2013;6:37. [Crossref] [PubMed]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38. [Crossref] [PubMed]
- Sugihara H, Ishimoto T, Miyake K, et al. Noncoding RNA Expression Aberration Is Associated with Cancer Progression and Is a Potential Biomarker in Esophageal Squamous Cell Carcinoma. Int J Mol Sci 2015;16:27824-34. [Crossref] [PubMed]
- Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog 2013;52:908-15. [Crossref] [PubMed]
- Ge XS, Ma HJ, Zheng XH, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci 2013;104:1675-82. [Crossref] [PubMed]
- Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol 2014;7:7938-44. [PubMed]
- Xie HW, Wu QQ, Zhu B, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol 2014;35:7743-54. [Crossref] [PubMed]
- Ohashi S, Miyamoto S, Kikuchi O, et al. Recent Advances From Basic and Clinical Studies of Esophageal Squamous Cell Carcinoma. Gastroenterology 2015;149:1700-15. [Crossref] [PubMed]
- Li X, Wu Z, Mei Q, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer 2013;109:2266-78. [Crossref] [PubMed]
- Lv XB, Lian GY, Wang HR, et al. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One 2013;8:e63516. [Crossref] [PubMed]
- Cao X, Zhao R, Chen Q, et al. MALAT1 might be a predictive marker of poor prognosis in patients who underwent radical resection of middle thoracic esophageal squamous cell carcinoma. Cancer Biomark 2015;15:717-23. [Crossref] [PubMed]
- Wang YL, Bai Y, Yao WJ, et al. Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous cell carcinoma and its correlation with tumor progression and patient survival. Int J Clin Exp Pathol 2015;8:11871-6. [PubMed]
- Shi WH, Wu QQ, Li SQ, et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol 2015;36:2501-7. [Crossref] [PubMed]
- Tong YS, Zhou XL, Wang XW, et al. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med 2014;12:233. [Crossref] [PubMed]
- Tong YS, Wang XW, Zhou XL, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer 2015;14:3. [Crossref] [PubMed]
- Zhou XL, Wang WW, Zhu WG, et al. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol Carcinog 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Yu X, Li Z. Long non-coding RNA HOTAIR: A novel oncogene Mol Med Rep 2015;12:5611-8. (Review). [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Zhang X, Zhou L, Fu G, et al. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis 2014;35:2062-7. [Crossref] [PubMed]
- Deng Q, Sun H, He B, et al. Prognostic value of long non-coding RNA HOTAIR in various cancers. PLoS One 2014;9:e110059. [Crossref] [PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. [Crossref] [PubMed]
- Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 2015;34:7. [Crossref] [PubMed]
- Yao W, Bai Y, Li Y, et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol 2016;37:4305-12. [Crossref] [PubMed]
- Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem 2015;290:3925-35. [Crossref] [PubMed]
- Wang W, Zhu Y, Li S, et al. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting beta-catenin via Ezh2. Oncotarget 2016. [Epub ahead of print]. [PubMed]
- Ling H, Spizzo R, Atlasi Y, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res 2013;23:1446-61. [Crossref] [PubMed]
- Cai Y, He J, Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther 2015;8:2657-64. [PubMed]
- Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol 2014;35:5375-80. [Crossref] [PubMed]
- Wang CY, Hua L, Yao KH, et al. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol 2015;8:779-85. [PubMed]
- Wang J, Qiu M, Xu Y, et al. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol 2015;36:5523-8. [Crossref] [PubMed]
- Cui F, Wu D, He X, et al. Long noncoding RNA SPRY4-IT1 promotes esophageal squamous cell carcinoma cell proliferation, invasion, and epithelial-mesenchymal transition. Tumour Biol 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Pasic I, Shlien A, Durbin AD, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res 2010;70:160-71. [Crossref] [PubMed]
- Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res 2013;41:4976-87. [Crossref] [PubMed]
- Wang PL, Liu B, Xia Y, et al. Long non-coding RNA-Low Expression in Tumor inhibits the invasion and metastasis of esophageal squamous cell carcinoma by regulating p53 expression. Mol Med Rep 2016;13:3074-82. [PubMed]
- Li J, Chen Z, Tian L, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014;63:1700-10. [Crossref] [PubMed]
- Pan F, Yao J, Chen Y, et al. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol 2014;7:2838-49. [PubMed]
- Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 2014;63:881-90. [Crossref] [PubMed]


