Global DNA methylation and PTEN hypermethylation alterations in lung tissues from human silicosis
Introduction
Silicosis is an occupational fibrotic lung disease caused by long-term inhalation of free crystalline silicon dioxide, or silica, which is an occupational health problem worldwide (1). In 1997, the International Agency for Research on Cancer (IARC) announced crystalline silica is carcinogenic to humans (group 1) (2). Silica dust can also increase the mortality rate from respiratory diseases and cardiovascular diseases (3). Exposure to silica can cause abnormal gene expression (4). Regulation of gene expression not only depends on genetic mechanism, also with epigenetics mechanisms without genetic coding change, including DNA methylation and histone acetylation and so on (5).
The characteristic pathological change forms silicotic nodules and diffuses interstitial fibrosis. Mitogen-activated protein kinases (MAPK) possesses complicated functions, for example extracellular regulated protein kinases (ERK) promotes cell proliferation, p38 MAPK and c-Jun N-terminal kinase (JNK) mediates cell death or cell survival depending on the stimuli (6-8). Phosphatidylinositol 3-kinase (PI3K) can activate protein kinase C (PKC) directly or indirectly through Serine/threonine kinase (AKT) activation, which subsequently results in ERK activation, which can be negatively modulated by phosphatase and tensin homolog deleted on chromosome 10 (PTEN) (9-12). Exposure to silica induced α-SMA expression via PI3K/AKT pathway, DNA double strand breaks, cyclooxygenase-2 expression via MAPKs and cell cycle changes via PI3K/AP-1 pathway in vitro (13-16).
Epigenetic regulation of gene expression has been widely studied in cancer (17,18), and aberrant DNA methylation plays a role in the development of various diseases. But the reports about the relationship between silica and aberrant DNA methylation have been limited and focus on certain gene, rats model and blood from silicosis patient (19-21). PTEN was downstream of PI3K using siRNA in silica-induced human embryo lung fibroblasts (HELFs) (unpublished data). It has been documented that PTEN promoter methylation mediated the loss of its expression implicated in hepatic stellate cell (22). The loss of PTEN function contributes to silica-mediated PI3K/AKT/MAPK/AP-1 pathway activation.
Taken together, we performed genome-scale DNA methyaltion profile of lung tissues from silicosis patients to identify DNA methyaltion patterns in silicosis through llumina Human Methylation 450K Beadchip (450K BeadChip). By screening the genes in differentiated CpG sites promoter between early-stage silicosis and advanced stage, immunohischemistry was performed to measure the level of proteins in these specimens and these gene methyaltion status was verified by methylation specific PCR (MS-PCR) in HELFs.
Methods
Reagents
RPMI 1640 medium was obtained from Thermo Fisher Scientific, USA. Fetal bovine serum (FBS) was purchased from Gibco, USA. L-glutamine and gentamycin sulfate were obtained from Sigma, USA.
Genome-wide DNA methylation analysis
Ten formalin-fixed, paraffin-embedded (FFPE) sections from silicosis patients were obtained from National Institute for Occupational Health and Poison Control, China. We selected patients with silicosis who had undergone autopsying between 1967 and 1979, and diagnosed lung cancer cases were excluded. The patients we selected in the paper had no other illness in the lung. And they were died because of the silicosis. We divided these samples based on disease progress, early stage or advanced stage. The first group included six samples, and the second group contained four samples. Normal lung tissues methylation data were obtained from GEO database. Genomic DNA was extracted from FFPE using QIAamp DNA FFPE Tissue Kit (Qiagen). Genomic DNA was bisulfite-converted using EZ DNA Methylation Kit (Zymo Research). Then the converted DNA was amplified at 37 °C for 22 h, fragmented, purified, resuspended and hybridized with multiBeadChip at 48 °C for 16 h. After which, the BeadChip was experienced to wash, extend the primers hybridized to the DNA by adding labeled nucleotides, and stained. The BeadChip was coated and scanned using the Illumina® iScan system. The image data was processing with the Genome StudioTM Methylation Module software and analyzed by Illumina Methylation Analyzer.
Immunohistochemistry
The above autopsy specimens and two normal lung tissues was measured 3 cm × 2 cm × 1 cm, and paraffin embedded and section was observed with hematoxylin and eosin (H&E) staining. Immunohistochemistry was performed to evaluate the levels of the c-Jun and PTEN protein.
Cell culture and silica exposure
HELFs were purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. HELFs were cultured in RPMI-1640 medium with 10% heat-inactivated FBS, 2.5 mmol/L glutamine, 100 µg/mL gentamycin sulfate at 37 °C in humidified atmosphere of 5% CO2. The silica particles were suspended in D-Hanks buffer saline, autoclaved to sterilize, and diluted to the needed concentrations (1 mg/mL).
MS-PCR
Genomic DNA of HELFs was extracted using Wizard® Genomic DNA Purification Kit (Promega, USA), according to the manufacturer’s instructions. The methylation status of the c-Jun and PTEN promoter region was detected by Methylation-Specific Polymerase Chain Reaction Genomic (MS-PCR). DNA was treated with sodium bisulfite using an EpiTect Bisulfite Kit (Qiagen, Germany). Two micrograms of DNA were modified in a final volume of 140 µL following the instructions of the manufactures. After bisulfite modifications, the MS-PCR for PTEN and c-Jun were conducted using ZymoTaq™ PreMix (Zymo Research, USA). The primers for the unmethylated PTNE gene were 5'-TATTAGTTTGGGGATTTTTTTTTTGT-3' (sense) and 5'-CCCAACCCTTCCTACACCACA-3' (antisense); the primers for the methylated PTEN gene were 5'-GTTTGGGGATTTTTTTTTCGC-3' (sense) and 5'-AACCCTTCCTACGCCGCG-3' (antisense) (23). Primers for c-Jun gene was designed by Methprimer (24), primer sequences were as follows: unmethylated reaction, sense primer, 5'-GGTAGTGGAGTATTATTTTATTTTGT-3', antisense primer, 5'-CAAAACCTTCCCATTAACTCAC-3'; methylated reaction, sense primer, 5'-GGGTAGCGGAGTATTATTTTATTTC-3', antisense primer, 5'-CAAAACCTTCCCATTAACTCG-3'. PCR was performed as follows: one cycle of 95 °C, and 40 cycles of 94 °C for 30 s, 60 °C (for detection of PTEN gene) or 56.6 °C (for detection of c-Jun gene) for 1 min and 72 °C for 1 min, followed by final extension at 72 °C for 5 min. Each product was loaded onto 2.5% agarose gel with ethidium bromide and visualized under UV.
Results
Chest radiography
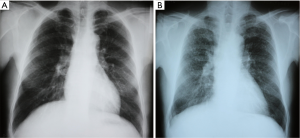
One of methods to diagnosis of silicosis is chest radiography. According to guidelines for the use of the International Labour Organization, the profusion of small opacities refers to the concentration of small opacities in affected zones of the lung. The category of profusion is based on comparisons with the standard radiographs. Classification of a radiograph using the 4-category and 12-subcategory scale is performed as Table 1. The appropriate category is chosen by comparing a subject radiograph with standard radiographs that define the levels of profusion characteristic of the centrally placed subcategories (0/0, 1/1, 2/2, 3/3) within these categories. Two kinds of shape of small opacities are recognized: rounded and irregular. In each case, three sizes are differentiated. For small rounded opacities, the three size ranges are denoted by the letters p, q and r. The three size ranges of small irregular opacities are denoted by the letters s, t and u. A large opacity is defined as an opacity having the longest dimension exceeding 10 mm. Categories of large opacities are defined as Table 1 (25).

Full table
We performed the chest radiography in early-stage group and advanced-stage group respectively. In early-stage group, rounded small opacities can be seen in upper and middle zones of both lung fields, mainly in lateral region, recognized as p/q. The profusion of small opacities is from 0/1 to 1/1 (Figure 1). In the advanced-stage group, there are increased and confused bronchovascular shadows in chest radiography. Rounded small opacities can be seen in lung fields, more in upper and middle zones of both lung fields, recognized as q/r. The profusion of small opacities is from 2/3 to 3/3 (Figure 1).
450K DNA methylation analysis of silicosis and normal lung tissues.
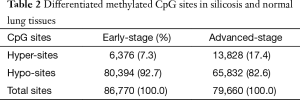
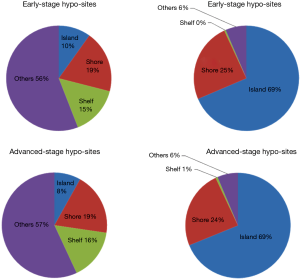
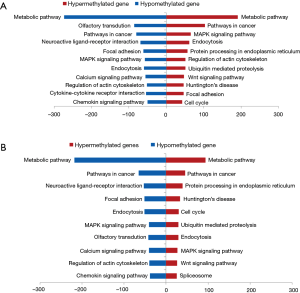
Methylation of 86,770 (18%) CpG sites out of 480,815 differed in early-stage and normal, which is 79,660 (16.6%) in advanced-stage. Hyper- or hypo-methylated CpG sites between silicosis and normal lung tissues were shown in Table 2. From these data, we concluded that the alteration of DNA methylation existed in silicosis lung tissues with hypomethylation of CpG sites being more common than hypermethylation. High methylated CpG sites are more likely to located in promoter regions either in early-stage or advanced stage. A total of 59.3% (3,780 sites) and 63.6% (8,797 sites) are associated with promoter regions, with hypomethylated CpG sites both only less than 2%, respectively. From the CpG content and neighborhood context, CpG sites significantly hypermethylated are in CpG island regions compared with normal lung tissue (Figure 2). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed to identify the biological process associated with the differentially methylated genes in early-stage silicosis or advanced-stage silicosis compared with normal lung tissue data. About 200 signaling pathways were detected and considered significant, according to the criterion of P≤0.001 and |difference score| ≥0.4 (Figure 3). Figure 4 showed the interaction in the differentially methylated gene of PI3K/AKT/MAPK/AP-1 cell signaling transduction pathway.
Full table
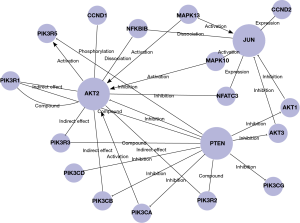
450K DNA methylation data on PI3K/PTEN/AKT/MAPK/AP-1cell signal pathway
To validate the effect of DNA methylation on this pathway, we select CpG promoter sites with these genes of PI3K/PTEN/AKT/MAPK/AP-1. MAPKs are family with several members, which can be hypermethylated or hypomethylated, so we not select MAPKs. Fos can not be screened under the criteria of P≤0.001 and |difference score| ≥0.4. Table 3 listed the genes under the above criteria. PTEN and Jun methylation were differentiated in disease stage, so their expression level was discussed in following study.
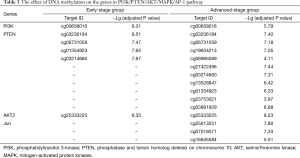
Full table
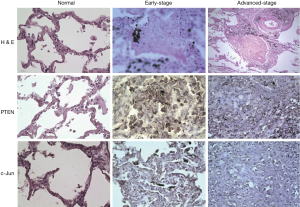
Immunhistochemical analyses of the level of PTEN and c-Jun in specimens from silicosis patients
Histological examination revealed many dust-laden macrophages around interlobular septa, and in peribronchiolar and perivascular areas, associated with some interstitial fibrosis and increased amounts of reticular fibers. The alveolar spaces were filled with swollen macrophages and amorphous proteinaceous semifluid. Those macrophages contained cytoplasmic black dust particles. There were some fibrotic nodules typical of pneumoconiosis (Figure 5, first row). The positive expression of c-Jun and PTEN protein are in early-stage cases and negative or mild expression in advanced-stage cases. All the cases were similar in c-Jun and PTEN protein expression which decreased relatively with the degree of pneumoconiosis lesions.
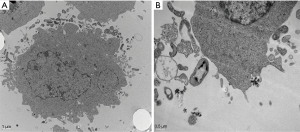
Measure the PTEN and c-Jun methylation status in HELFs
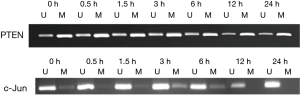
Electron microscopy showed that compared with control group, the experimental group exposure to 100 µg/mL silica suspension swallowed particulate in 24 h (Figure 6). In order to verify the above results, we selected HELF cells exposed to 100 µg/mL silica suspension. DNA was extracted after exposing 0.5, 1, 3, 6, 12 and 24 h. For PTEN, there are methylated bands and unmethylated bands at all-time points. However, c-Jun had shown differed methylation at 12 and 24 h, only having unmethylated bands (Figure 7).
Discussion
This is the first study to compare differential DNA methylation on a genome-scale level in lung tissues from silicosis patients with normal lung tissue methylation data. FFPE specimens from 6 early-stage silicosis cases and 4 advanced-stage silicosis cases showed altered DNA methylation in global scale, which were involved in varied signaling pathway. Compared with early-stage group, the level of PTEN and c-Jun protein, existed in PI3K/PTEN/AKT/AP-1 cell signaling pathway, decreased in advanced-stage group.
Aberrant DNA hypermethylation often occurs in the promoters associated CpG islands, the clusters of CpG dinucleotides, resulting in transcription silencing and vice versa (26). Epigenetic modifications are not only with cancer, more and more data supported that this may related with variable diseases. The pattern of DNA methylation is influenced by various environment factors (27), including silica particle. The 450K BeadChip has been reported as an accurate and reproducible tool to examine DNA methylation in genomic scale, which allows the identification of CpG sites location. It covers 96% of the CpG islands, associated with 56.1% of gene promotors, and 99% of the Ref Seqgenes (28-30). In our study, from the genomic wide, hypomethylated CpG sites seem to be more common, while hypermethylated CpG sites are more likely to link to the promoter regions (59.2% and 63.6%, respectively). KEGG analysis shows different pathway derived from high methylated genes and low methylated genes. Some of these differentially methylated pathways have known on the effect of inflammation and fibrosis.
Exposure to silica results in differences in gene expression, which can be a part accounted by DNA methylation. For example, DNA methylation in the promoter region of (ADP-ribose) polymerases-1 (PARP-1) might be responsible for silica-induced DNA double strand breaks in vitro (19,20). In addition, silica-induced tumors tissues exhibited widespread genomic hypomethylation in rat model (21). Furthermore, silicosis patients with lung cancer had higher risk of aberrant promoter methylation in at least one of the five tumor suppressor genes than those without lung cancer (31). The reports about the relationship between silicosis and DNA methylation have limited. IPF is a disease similarity with silicosis. Genome-wide methylation arrays show that 402 differentially methylated CpG islands overlapped between IPF and lung cancer, three genes expression positive correlation with hypomethylated promoters (32). The Human Methylation 27 array find that multiple CpG sites across the genome are differentially methylated in IPF fibroblasts (33). In our study, we choose FFPE lung tissues of silicosis without cancer, and we will explore the silicosis lung tissues with cancer and look forward their similarities and differences.
PI3K can activates PKC directly or indirectly through AKT activation, which subsequently results in ERK activation, which can be negatively modulated by PTEN (9-12). The decreased expression of PTEN may contribute to the activation of PI3K/AKT/MAPK/AP-1 cell signaling transduction pathway, which regulates the cell cycle (15). AP-1 is the dimers composed of c-Jun and c-Fos, which is an intersection of many signaling pathway within the cell nucleus. In addition, histone deacetylase regulated fibroblast-myofibroblast differentiation by phosphorylation of AKT (34). Therefore, the expression of signal molecular was not only related to the upstream, but also to epigenetic modulation. Hypermethylation is one of the mechanisms of PTEN inactivation, which has been widely reported in many types of cancer (23,35). PTEN methylation at the promoter region is responsible for the down-regulation of its expression (22), accompanied with activation of PI3K/AKT/mTOR pathway (36). AKT can stabilize and enhance the nuclear translocation of DNA methyltransferase 1 (DNMT1) (37,38), and DNMT1 contributes to the hypermethylation of PTEN (22). c-Jun is an important member of the activator-protein 1 (AP-1) complex, regulating cellular survival or proliferation by interacting with specific target DNA sequence to regulate gene expression (39). c-Jun is regulated not only by MAPK pathway, but also by dynamic acetylation of lysine 4-methylated histone H3 or acetylation of phosphorylated histone H3 (40,41). So, DNA methylation was not the exclusive way for regulation of c-Jun.
In our study, we chose silicosis specimens without lung cancer. Silicosis patients have high risk to develop lung cancer. Although fibrosis may be a so-called precancerous lesion (42), DNA methylation patterns seems to have some differences between fibrosis and cancer. Hypomethylation of CpG sites is more frequently observed than hypermethylation in cancer and lesions from epithelial hyperplasia to lung cancer accompanied with decreased DNA methylation (18,43). Moreover, LINE-1 retrotransposon differs in methylation status of IPF and lung cancer (32). So, our next research will select tissues from silicosis patients with lung cancer to disclose their similarities and differences in DNA methylation.
Research on target organs lesions has profundity and extent for us to understand pathological process. As with other studies, silicosis lung tissues have heterogeneity. The silicosis lung tissues contain epithelial, endothelial cells, fibroblasts, and many inflammatory cells. Naturally, it is possible to underestimate DNA methylation and not to identify DNA methylation related to specific cell type. Another limitation of our study is normal lung tissues methylation data obtained from CEO, which is not an age-matched sample with silicosis patients.
Conclusions
These results suggested that abnormal DNA methylation on genome-scale was implicated in silicosis, and PTEN promoter hypermethylation might be associated with the decrease of PTEN protein.
Acknowledgements
Funding: This study was supported by National Nature and Science Foundation of China (grant No. 81472956 and 30972449) and the National Key Technology Research and Development Program (2014BAI12B02).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the medical ethics committee of National Institute for Occupational Health and Poison Control, Chinese Center for Disease Control and Prevention (No. 201402) and written informed consent was obtained from all patients.
References
- WHO. The Global Occupational Health Network: elimination of silicosis. Available online: http://www.who.int/occupational_health/publications/newsletter/gohnet12e.pdf
- Cancer IAfRo. IARC Monographs on the evaluation of carcinogenic risks to humans, vol 68: silica, some silicates, coal dust and para-aramid fibrils. Lyon: International Agency for Research on Cancer, 1997.
- Chen W, Liu Y, Wang H, et al. Long-term exposure to silica dust and risk of total and cause-specific mortality in Chinese workers: a cohort study. PLoS Med 2012;9:e1001206. [Crossref] [PubMed]
- Perkins TN, Shukla A, Peeters PM, et al. Differences in gene expression and cytokine production by crystalline vs. amorphous silica in human lung epithelial cells. Part Fibre Toxicol 2012;9:6. [Crossref] [PubMed]
- Liu L, Li Y, Tollefsbol TO. Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol 2008;10:25-36. [PubMed]
- Thornton TM, Rincon M. Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int J Biol Sci 2009;5:44-51. [Crossref] [PubMed]
- Xia Z, Dickens M, Raingeaud J, et al. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 1995;270:1326-31. [Crossref] [PubMed]
- Kostadinova R, Montagner A, Gouranton E, et al. GW501516-activated PPARβ/δ promotes liver fibrosis via p38-JNK MAPK-induced hepatic stellate cell proliferation. Cell Biosci 2012;2:34. [Crossref] [PubMed]
- Toker A, Meyer M, Reddy KK, et al. Activation of protein kinase C family members by the novel polyphosphoinositides PtdIns-3,4-P2 and PtdIns-3,4,5-P3. J Biol Chem 1994;269:32358-67. [PubMed]
- Le Good JA, Ziegler WH, Parekh DB, et al. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 1998;281:2042-5. [Crossref] [PubMed]
- Schönwasser DC, Marais RM, Marshall CJ, et al. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol 1998;18:790-8. [Crossref] [PubMed]
- Chetram MA, Hinton CV. PTEN regulation of ERK1/2 signaling in cancer. J Recept Signal Transduct Res 2012;32:190-5. [Crossref] [PubMed]
- Li AP, Hou ZG, Fan JJ, et al. Silica induced α-SMA expression in HBE cell line by targeting the PI3K/Akt pathway. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 2012;30:937-40. [PubMed]
- Msiska Z, Pacurari M, Mishra A, et al. DNA double-strand breaks by asbestos, silica, and titanium dioxide: possible biomarker of carcinogenic potential? Am J Respir Cell Mol Biol 2010;43:210-9. [Crossref] [PubMed]
- Jia X, Liu B, Shi X, et al. Roles of the ERK, JNK/AP-1/cyclin D1-CDK4 pathway in silica-induced cell cycle changes in human embryo lung fibroblast cells. Cell Biol Int 2011;35:697-704. [Crossref] [PubMed]
- Tomaru M, Matsuoka M. The role of mitogen-activated protein kinases in crystalline silica-induced cyclooxygenase-2 expression in A549 human lung epithelial cells. Toxicol Mech Methods 2011;21:513-9. [Crossref] [PubMed]
- Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012;22:271-82. [Crossref] [PubMed]
- Shen J, Wang S, Zhang YJ, et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics 2013;8:34-43. [Crossref] [PubMed]
- Gong C, Tao G, Yang L, et al. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol Lett 2012;209:264-9. [Crossref] [PubMed]
- Liu HF, Zhang FM, Liu BC, et al. Roles of DNA dependent protein kinase in silica-induced cyclin E and CDK2 expressions and cell cycle changes in human embryo lung fibroblasts. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi 2011;29:241-5. [PubMed]
- Blanco D, Vicent S, Elizegi E, et al. Altered expression of adhesion molecules and epithelial-mesenchymal transition in silica-induced rat lung carcinogenesis. Lab Invest 2004;84:999-1012. [Crossref] [PubMed]
- Bian EB, Huang C, Ma TT, et al. DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell activation and liver fibrogenesis in rats. Toxicol Appl Pharmacol 2012;264:13-22. [Crossref] [PubMed]
- Soria JC, Lee HY, Lee JI, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res 2002;8:1178-84. [PubMed]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427-31. [Crossref] [PubMed]
- ILO. Guidelines for the use of the ILO International Classification of Radiographs of Pneumoconioses, Revised edition 2011. Occupational Safety and Health Series No. 22 (Rev.2011). Geneva: International Labour Office, 2011.
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [PubMed]
- Yang IV, Schwartz DA. Epigenetic control of gene expression in the lung. Am J Respir Crit Care Med 2011;183:1295-301. [Crossref] [PubMed]
- Dedeurwaerder S, Defrance M, Calonne E, et al. Evaluation of the Infinium Methylation 450K technology. Epigenomics 2011;3:771-84. [Crossref] [PubMed]
- Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011;6:692-702. [Crossref] [PubMed]
- Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 2004;20:1170-7. [Crossref] [PubMed]
- Umemura S, Fujimoto N, Hiraki A, et al. Aberrant promoter hypermethylation in serum DNA from patients with silicosis. Carcinogenesis 2008;29:1845-9. [Crossref] [PubMed]
- Rabinovich EI, Kapetanaki MG, Steinfeld I, et al. Global methylation patterns in idiopathic pulmonary fibrosis. PLoS One 2012;7:e33770. [Crossref] [PubMed]
- Huang SK, Scruggs AM, McEachin RC, et al. Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung. PLoS One 2014;9:e107055. [Crossref] [PubMed]
- Guo W, Shan B, Klingsberg RC, et al. Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol 2009;297:L864-70. [Crossref] [PubMed]
- Rizvi MM, Alam MS, Ali A, et al. Aberrant promoter methylation and inactivation of PTEN gene in cervical carcinoma from Indian population. J Cancer Res Clin Oncol 2011;137:1255-62. [Crossref] [PubMed]
- Mueller S, Phillips J, Onar-Thomas A, et al. PTEN promoter methylation and activation of the PI3K/Akt/mTOR pathway in pediatric gliomas and influence on clinical outcome. Neuro Oncol 2012;14:1146-52. [Crossref] [PubMed]
- Sun L, Zhao H, Xu Z, et al. Phosphatidylinositol 3-kinase/protein kinase B pathway stabilizes DNA methyltransferase I protein and maintains DNA methylation. Cell Signal 2007;19:2255-63. [Crossref] [PubMed]
- Hodge DR, Cho E, Copeland TD, et al. IL-6 enhances the nuclear translocation of DNA cytosine-5-methyltransferase 1 (DNMT1) via phosphorylation of the nuclear localization sequence by the AKT kinase. Cancer Genomics Proteomics 2007;4:387-98. [PubMed]
- Leppä S, Bohmann D. Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene 1999;18:6158-62. [Crossref] [PubMed]
- Hazzalin CA, Mahadevan LC. Dynamic acetylation of all lysine 4-methylated histone H3 in the mouse nucleus: analysis at c-fos and c-jun. PLoS Biol 2005;3:e393. [Crossref] [PubMed]
- Clayton AL, Rose S, Barratt MJ, et al. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J 2000;19:3714-26. [Crossref] [PubMed]
- Vancheri C, Failla M, Crimi N, et al. Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur Respir J 2010;35:496-504. [Crossref] [PubMed]
- Piyathilake CJ, Frost AR, Bell WC, et al. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol 2001;32:856-62. [Crossref] [PubMed]


