The use of endobronchial ultrasound guided transbronchial needle aspiration specimens for next generation sequencing in non-small cell lung cancer: a clinical perspective
Introduction
The National Comprehensive Cancer Network (NCCN) 2017 Clinical Practice Guidelines for Non-Small Cell Lung Cancer (NSCLC) recommend utilization of a procedure that allows concurrent diagnosis, staging and procurement of material, which is sufficient for genetic testing (1). As such guidelines recommend endobronchial ultrasound (EBUS) guided transbronchial needle aspiration (TBNA) as the principal test for the diagnosis and staging of lung cancer (2). The majority of patients with lung cancer continue to be diagnosed at an advanced stage for which broad molecular profiling is recommended and, in an era in which small volume small volume biopsy or cytology specimens have become the main diagnosis modality for most patients with lung cancer, it would be advantageous to be able to utilize these small specimens for molecular testing (1,3).
The recommendations from the NCCN guidelines for broad molecular profiling go beyond testing solely for EGFR, ALK, and ROS-1. While there are different approaches to broad molecular profiling, next generation sequencing (NGS) is a method which utilizes a single test to identify thousands of somatic or germline mutations from hundreds of genes. In fact, this process allows the entire cancer genome and transcriptome to be examined (4). As a result, NGS profiling is able to identify currently targetable mutations or other genetic abnormalities which may be eligible for current or future clinical trials. In 26% of patients with NSCLC, NGS reveals mutations for which a targeted agent is available and recommended by NCCN guidelines. Another 39% of patients tested by NGS are discovered to harbor a genomic alteration for which a targeted agent is available on a clinic trial (5). In addition, when compared to conventional testing, NGS has been shown to be more sensitive for identifying these targetable or potentially targetable driver-mutations (e.g., EGFR, KRAS, ALK, ROS1, RET) (5,6). Along with broad molecular testing at the time of diagnosis, it is advised that patients undergo comprehensive genetic profiling when disease progression is noted in order to evaluate for secondary mutations; this may help guide referrals towards clinical trials and offer prognostic value (1,7,8). As an example, NGS testing allows for the identification and targeting of mechanisms of resistance to (I) EGFR TKIs (T790 M mutation, MET amplification, BRAF and PIK3CA mutations) and (II) first generation ALK inhibitors (crizotinib). This will allow further treatment with osimertinib or newer ALK inhibitors (e.g., ceritinib, alectinib), respectively (7,9-11). Complex resistance mechanisms are only discovered by testing broad gene panels, which is feasible with NGS (11).
Although there are studies published on the suitableness of formalin fixed paraffin embedded (FFPE) cell block samples for NGS testing (12,13), cytology smear preservation methods provide a superior quality DNA for testing (14-16). Moreover, an additional advantage of cytology slides lies in the ability for rapid onsite examination (ROSE) to assess smears for cellularity and adequacy in real time in the course of the procedure. The adequacy of a cell block cannot be reliably assessed intra-procedurally. The importance of adequate sample cellularity is highlighted by prior studies which describe the successful technique of extracting DNA from cytology slides prepared with fine needle aspiration (FNA) tumor samples (17-21). These studies demonstrate a direct relationship between successful NGS testing and increased slide cellularity. NGS sequencing of 50 target genes was successful greater 95% of the time on slides with more than 5 ng/mL of DNA, which equates to a slide with at least 5,000 cells (17). Recent publications reporting on NGS panels of greater than 200 genes are remarkable for all employing histology or cell block samples; there are no reports of these large panels being run on smears (5,13).
In this viewpoint article we describe the workflow which is practiced at the University of Chicago Medical Center for NGS testing of 50 gene (Oncoscreen) and greater than 1,000 gene (OncoPlus) panels on slide cytology specimens procured by EBUS-TBNA sampling of metastatic interlobar, hilar, or mediastinal lymph nodes in patients with NSCLC.
Technique: collection, handling and processing of specimens
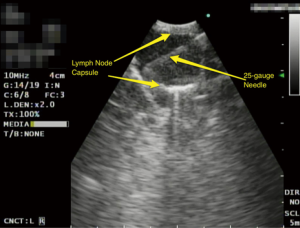
A standardized examination of the interlobar, hilar and mediastinal lymph nodes is employed. If EBUS identifies a lymph node greater than 5 mm, then a 25-gauge needle EBUS TBNA of the lymph node is performed (Figure 1). A routine EBUS-TBNA procedure results in the acquisition of at least three aspirates from each lymph node and this process is carried forth systematically through all remaining lymph node stations to be sampled.
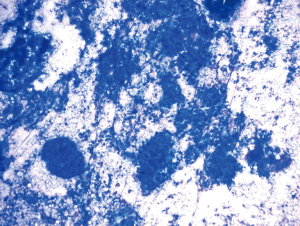
Subsequent to each sample aspiration, the biopsy needle is removed from the bronchoscope and a single drop of sample material is deposited onto a labeled glass slide. The sample is delivered first by advancing the stylet through the needle channel and, if it results in an insufficient discharge of material, this is followed by the attachment of an empty 10 cc syringe to the stylet hub and flushing of air through the channel. After the deposition of a small amount of material onto a slide the remaining aspirate in the needle from each pass is expelled into formalin or Cytolyt solution and is subsequently processed into a cell-block. After material is expelled onto the glass slide, a second slide is used to smear the sample resulting in the creation of two slides. Diff-Quik is used to stain one of the slides which is then immediately evaluated by an onsite cytopathologist (Figure 2). The second smear is preserved for future Pap staining by sprayed-fixing it with alcohol.
If the cytopathologist favors a diagnosis of non-small cell lung cancer, then the smear cellularity is further evaluated for adequacy for downstream molecular studies. The estimation of cellularity is guided by the experience of the cytopathologist. At the University of Chicago Medical Center, we have found that the Oncoscreen panel requires approximately 2,000 tumor cells and that for the OncoPlus panel 20,000 tumor cells are necessary. In the experience of our institution, a smear with greater than 50% tumor will have a sufficient cellularity for both panels. On ROSE, if a smear is interpreted as sufficient for a diagnosis but is considered to have a cellularity insubstantial for molecular studies, the cytopathologist will request additional passes which continue to be performed and evaluated until an adequate smear is obtained. If necessary, a combination of several smears may be used to surpass the minimum cellularity requirement.
NGS techniques
Fusion panel—Genes analyzed for fusions/translocations: ALK, RET, and ROS1. A full description is provided online (http://uchicagomedlabs.testcatalog.org/show/FUSNCS)
OncoScreen—Solid tumor mutation testing by NGS for 50 genes that include all targetable mutations with drugs approved by NCCN guidelines. A full description and list of covered genomic regions within these genes is provided online (http://uchicagomedlabs.testcatalog.org/show/NGFFPE-FFPE-NGSCS-Cytology-Smear--1)
OncoPlus—Somatic mutation testing by NGS that is designed to interrogate 1,213 cancer-related genes with a subset clinically reported for personalized care. A full description and list of covered genomic regions within these genes is provided online (http://uchicagomedlabs.testcatalog.org/show/NGPLSC)
Conclusions
Currently approved drugs and those in clinical trials are being used to successfully target driver mutations in NSCLC. The guidelines have been updated to reflect this trend and recommendations for broad molecular profiling for NSCLC is generally accepted for the individualized treatment of lung cancer. EBUS-TBNA represents a minimally invasive procedure which allows for immediate assessment of adequacy for genetic testing via ROSE. In the current era, such EBUS-TBNA specimens are most likely to be the first, and may indeed be the only, source of material obtained during the course of disease in patients suffering from lung cancer. It is promising that this procedure can provide samples for comprehensive genetic testing which have optimal DNA quality (e.g., cytology smears) and may therefore allow personalized treatment at time of diagnosis or disease progression without the requirement for additional tissue acquisition by invasive procedures.
Acknowledgements
None.
Footnote
Conflicts of Interest: Dr Murgu is an educational consultant for Olympus America, Cook Inc., Boston Scientific and Medtronic. The other author has no conflicts of interest to declare.
References
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Non-Small Cell Lung Cancer. Version 3.2017 — November 16, 2016. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- Silvestri GA, Gonzalez AV, Jantz MA, et al. Methods for staging non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e211S-50S.
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Blumenthal GM, Mansfield E, Pazdur R. Next-Generation Sequencing in Oncology in the Era of Precision Medicine. JAMA Oncol 2016;2:13-4. [Crossref] [PubMed]
- Drilon A, Wang L, Arcila ME, et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin Cancer Res 2015;21:3631-9. [Crossref] [PubMed]
- Lim SM, Kim EY, Kim HR, et al. Genomic profiling of lung adenocarcinoma patients reveals therapeutic targets and confers clinical benefit when standard molecular testing is negative. Oncotarget 2016;7:24172-8. [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Novello S, Barlesi F, Califano R, et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-v27. [Crossref] [PubMed]
- Kim H, Chung JH. Overview of clinicopathologic features of ALK-rearranged lung adenocarcinoma and current diagnostic testing for ALK rearrangement. Transl Lung Cancer Res 2015;4:149-55. [PubMed]
- Bosc C, Ferretti GR, Cadranel J, et al. Rebiopsy during disease progression in patients treated by TKI for oncogene-addicted NSCLC. Target Oncol 2015;10:247-53. [Crossref] [PubMed]
- Sullivan I, Planchard D. ALK inhibitors in non-small cell lung cancer: the latest evidence and developments. Ther Adv Med Oncol 2016;8:32-47. [PubMed]
- Qiu T, Guo H, Zhao H, et al. Next-generation sequencing for molecular diagnosis of lung adenocarcinoma specimens obtained by fine needle aspiration cytology. Sci Rep 2015;5:11317. [Crossref] [PubMed]
- Young G, Wang K, He J, et al. Clinical next-generation sequencing successfully applied to fine-needle aspirations of pulmonary and pancreatic neoplasms. Cancer Cytopathol 2013;121:688-94. [Crossref] [PubMed]
- Fischer AH, Cibas ES, Howell LP, et al. Role of cytology in the management of non-small-cell lung cancer. J Clin Oncol 2011;29:3331-2. [Crossref] [PubMed]
- Vincek V, Nassiri M, Nadji M, et al. A tissue fixative that protects macromolecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab Invest 2003;83:1427-35. [Crossref] [PubMed]
- Treece AL, Montgomery ND, Patel NM, et al. FNA smears as a potential source of DNA for targeted next-generation sequencing of lung adenocarcinomas. Cancer Cytopathol 2016;124:406-14. [Crossref] [PubMed]
- Gleeson FC, Kipp BR, Levy MJ, et al. Lung cancer adrenal gland metastasis: Optimal fine-needle aspirate and touch preparation smear cellularity characteristics for successful theranostic next-generation sequencing. Cancer Cytopathol 2014;122:822-32. [Crossref] [PubMed]
- Gleeson FC, Kipp BR, Kerr SE, et al. Characterization of endoscopic ultrasound fine-needle aspiration cytology by targeted next-generation sequencing and theranostic potential. Clin Gastroenterol Hepatol 2015;13:37-41. [Crossref] [PubMed]
- Karnes HE, Duncavage EJ, Bernadt CT. Targeted next-generation sequencing using fine-needle aspirates from adenocarcinomas of the lung. Cancer Cytopathol 2014;122:104-13. [Crossref] [PubMed]
- Kanagal-Shamanna R, Portier BP, Singh RR, et al. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol 2014;27:314-27. [Crossref] [PubMed]
- Scarpa A, Sikora K, Fassan M, et al. Molecular typing of lung adenocarcinoma on cytological samples using a multigene next generation sequencing panel. PLoS One 2013;8:e80478. [Crossref] [PubMed]


