Influenza A induced cellular signal transduction pathways
Influenza A transmission
The influenza A virus results in 3 to 5 million severe cases globally each year. In the United States the Centre for Disease Control estimated over a 30 year period from 1976-1977 season to 2006-2007 season there were an average number of deaths of 23,607 per year. Interestingly the range of influenza associated deaths over this period varied from a low of 3,349 to a high of 48,614 deaths. This highlights the variable nature and pathogenicity of different influenza strains (1). The authors of this report also noted that seasons where H3N2 was prominent, the deaths associated with influenza was 2.7 times higher than in seasons where other strains were prominent. The variability of different strains also plays a role not only in the pathogenicity of influenza but also in its ability to be transmitted from person to person (1). The influenza A virus is responsible for seasonal epidemics and infrequently global pandemics of the flu as was the case with pandemic H1N1 in 2009. Seasonal outbreaks of the flu are a result of continual mutation of this virus with regards to the proteins recognized by the immune system while pandemic strains result from antigenic shifts which completely change the viral proteins the immune system responds to. H1N1 is estimated to be responsible for approximately between 151,700 and 575,400 deaths globally (2). Although the risk factors for development of severe complication from the flu were similar between seasonal and H1N1 infections for individuals with chronic lung diseases, pre-existing cardiac disease and pregnancy they differed in that obesity and people of younger ages cohorts (younger than 65) developed greater complications with H1N1 then those with seasonal strains (3).
The influenza virus is transmitted by three main modes which are (I) direct contact with an infected fomite (II) inhalation of droplets and (III) small particle aerosols. Infected droplets are spread primarily through coughing while aerosols are created during sneezing, speaking or breathing (4). Infected aerosols with particle sizes below 5 µm have been shown to remain aerosolized for 1 hour (5). To spread from one person to the next the influenza virus must remain viable while in the aerosol or on inanimate objects (6). Viability of influenza outside of the body has been studied and factors affecting the viability of this virus have been identified. Relative humidity is one factor studied. The highest viability was associated with humidity levels close to 100% which mimics the levels within the respiratory system (7). Infectious aerosols released in the air where the relative humidity ranged from 50% to 84% displayed a steady decrease in viability. When the humidity level dropped below 50% viability of the influenza virus increased (7). The variation of viability was associated with aerosol dehydration caused by relative humidity levels which increased the salt concentration to a threshold range where the protein content of respiratory aerosols protected the virus from further decline in viability (8). The changes in humidity are also associated with the different seasonal activities of influenza over different regions of the globe. Temperate climate regions generally experience high influenza activity during the times of lowest humidity and coldest temperatures while tropical regions experience influenza peaks during their rainy season where the humidity levels are close to 100%. In both regions the tendency of people to congregate indoors also corresponds to this timing (9).
Infection with influenza A virus results in developing the flu. The symptoms of a typical uncomplicated influenza infection are fever, chills, rhinorrhea, nasal congestion, sinus pain, sore throat, cough, headache, anorexia and myalgia (10). The influenza virus generally does not directly cause these symptoms but triggers an immune response that causes their development (11). Macrophages continually monitor the epithelium of the airways. In the presence of the influenza virus macrophages become activated and trigger the acute phase response. Release of chemokines from activated immune cells further recruits more immune cells to the infected tissue resulting in inflammation that triggers systemic symptoms associated with the flu (10). Fever is the most common symptom of the flu and is caused by the response to the release of cytokines including TNFα, IL6 and IL1 (11). These cytokines whether they are produced locally in the lung or systemically produce the systemic effect of fever. These cytokines cross the blood brain barrier reaching the central nervous system. There they interact with the vagus nerve and signal to the temperature control area of the hypothalamus. This initiates signaling pathways that results in the hypothalamus releasing mediators that cause reflex shivering, peripheral blood vessel constriction that results in the sensation of chills. The sensation of sore throat is attributed to the production of bradykinins and prostaglandins in the airways (10).
Influenza A viral structure
The influenza A virus is a negative sense segmented RNA virus that contains a lipid envelope (Figure 1). The core of the virus particle contains 8 segments of viral RNA that are coated by a viral protein nucleoprotein (NP) (12,13). The protein coated segments are also bound to the three protein subunits polymerase basic 1, polymerase basic 2 and polymerase acidic (PB1, PB2, PA) that comprise the viral RNA dependent RNA polymerase which is responsible for both transcription and replication of the viral genome (14). Together these are the components of the viral ribonucleoproteins (vRNP) which are surrounded by a protein layer made from the viral protein matrix protein 1 (M1) (15). This protein shell is covered by the envelope which is a lipid bilayer derived from the host cell membrane. The envelope contains three viral proteins hemagglutinin (HA), neuraminidase (NA) and matrix protein 2 (M2). The lipids of the envelope are enriched with sphingolipids and cholesterol forming rafts (16). These raft domains associate with the embedded envelope proteins HA, NA and M2. The stability of the virus particle is primarily governed by the lipids of the membrane. The envelope proteins HA and NA have a low mobility with in the envelope and reinforce the virus structure through there association with the underlying M1 protein. M1 links the vRNPs to the envelope glycoproteins contributing to an overall stabilizing effect of the virus structure (17).

The virus gains access to the body through the nose, mouth or eyes. Influenza A virus initiates an infection when it comes in contact with the mucosal surfaces of the respiratory tract (Figure 2). Influenza A must come in direct contact with the epithelial cells of the respiratory tract to initiate an infection. Here the virus encounters its first barrier, the layer of mucus that covers and protects the epithelial cells of the respiratory tract. The envelope glycoprotein NA cleaves sialic acid from mucin that lines the epithelial cells. The virus attaches to the exposed cells by the second envelope glycoprotein HA. The HA protein forms a homotrimer in the envelope. The monomer of HA is composed of two polypeptides termed Ha1 and Ha2. Ha1 possess the receptor binding domain while Ha2 contains the membrane fusion peptide (18). This protein attaches to sialic acid linked glycoproteins on the surface of epithelial cells. The type of sialic acid linkage to the glycoprotein determines the tropism of the different types of influenza A viruses towards different hosts (19). Human influenza A viruses contains a HA which has a binding preference for an α-2,6 linked sialic acid to galactose where as the avian origin influenza HA has a binding preference for an α-2,3 linked sialic acid to galactose. The distribution of these glycosidic residues within a species also determines the tissue tropism of influenza. The human respiratory tract contains predominantly α-2,6 linkages expressed on the surface of the epithelium. HA also is a major determinant for tissue tropism of influenza (4). HA must be cleaved in order for it to be functional. Proteases found ubiquitously expressed in the lungs are able to cleave HA in to a functional viral protein. Strains of influenza possessing HA that can be cleaved by other classes of proteases are able to disseminate, infecting a larger range of tissues and tend to be more pathogenic (19). In addition to sialic acid binding recent studies have identified alternative modes of influenza A entry into cells that are dependent on c type lectins and independent of sialic acid binding (20). Other requirements for influenza A viral attachment are also being elucidated. Fibronectin an extracellular matrix protein has been identified as a requirement for certain strains of H1 and H3 influenza viruses. Binding of the virus to the cell surface induces uptake through endocytosis. This uptake has been shown to utilize clatherin dependent and caveolin dependent mechanisms (21). Once the virus is endocytosed it remains in the maturing endosome. Influenza escapes the endosome in a pH dependent manner (22). As the endosome matures it acidifies which activates the M2 envelope protein. The M2 protein is a tetramer integral protein that forms a proton selective ion channel. This protein contains three major domains, a short ectodomain, a transmembrane domain that forms the ion channel and a long cytoplasmic domain (23). Activation of M2 allows protons to enter the viral particle. At a critical pH the M1 protein that stabilizes and mediates attachment of the vRNP to the envelope proteins dissociates and undergoes a morphological change freeing the vRNP from the envelope. In addition the Ha2 peptide of the HA protein activates allowing for the fusion between the membrane of the endosome and the viral envelope. This fusion allows for the cytosolic release of the vRNP from the endosome (24). Once inside the cytoplasm the vRNP must enter the nucleus for transcription of viral genes and replication of the viral genome (25). Entry of the infecting vRNP is mediated by NP which possesses a nuclear localization sequence that binds with karyopherin which is an endogenous heterodimer of karyopherin α1 with karyopherin β or karyopherin α2 and karyopherin β. Once bound by karyopherin the vRNP is ferried to and docked at the nuclear pore complex. Entry in to the nucleus is Ran 10 and p10 dependent (26). Once in the nucleus transcription of the viral genes occurs. The mechanism of viral transcription and genome replication has been well studied but the regulation and switching from transcription to genome replication remains to be fully elucidated. Progress has been made with regards to identifying viral elements involved in regulating these processes. Umbach et al. have found that influenza A virus produces short RNA nucleotides ranging from 18 to 27 nucleotides in length. These viral leader RNAs were found to originate from each of the 8 segments 5' ends (27). Perez et al. found that these virally encoded RNAs acted as enhancers for viral replication but they did not function as a primer for replication. These short RNAs were retained in the nucleus throughout the infection and it is postulated that they act to not only stabilize viral RNA genome replication but to also play a role in maintaining the proper stoichiometry of each viral segment (28). Viral proteins also have been shown to regulate viral transcription and genome replication. Widjaja et al. produced a model whereby transcription of viral genes is accomplished by the associated RNA dependent RNA polymerase. Production and nuclear accumulation of new polymerase allows for complementary RNA to be made. This serves as a template for viral genome replication which is stabilized by both the newly synthesized polymerase and NP (29). They also identified elements of the 3' and 5' UTR of the viral segments involved in competition for viral proteins required for replication and also demonstrated that shorter viral segments can out compete larger segments for viral proteins (29). The viral genomic RNA is associated with viral NP and the three polymerase subunits before being exported out of the nucleus. This requires the re-import of translated NP and the polymerase subunits which posses multiple elements that allow for shuttling between the nucleus and cytoplasm (30). Together these proteins form the vRNP. The vRNP forms a complex in the nucleus between the M1 protein and the nuclear export protein (NEP) formerly known as NS2 (31). Both M1 and NEP have been shown to be required for export of the vRNP out of the nucleus into the cytoplasm which is chromosome region maintenance I (CRM1) dependent (31). The vRNPs accumulate at the microtubule organizing centre upon exiting the nucleus. In addition to these viral proteins, a host cell protein Y-box binding protein (YB-1) has been identified as associating with this complex in the nucleus. This protein is believed to act as an adaptor for the vRNP allowing it to associate with microtubules and to target it to the rab11a recycling endosome which delivers the vRNP to the site of virus assembly (32).
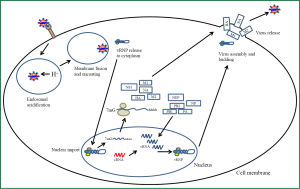
Release of the virus from the cells requires assembly of the viral proteins and all 8 vRNP segments into the viral particle. This assembly occurs at the cell’s plasma membrane. The viral proteins HA, NA and M2 are transmembrane proteins that are inserted into the host’s plasma membrane (33). HA and NA are localized to lipid rafts rich in cholesterol and sphingolipids (34). The M1 protein forms associations with the cytoplasmic portions of these transmembrane proteins. It has been demonstrated that viral assembly is selective for each of the 8 vRNPs and virions have one copy of each segment in them (13). Initiation of the budding process involves the HA protein which appears to be necessary for efficient budding but is not absolutely required. It has been shown that HA assists in creating membrane curvature to start the bud but requires the recruitment of other viral proteins for the budding process to complete. The M2 protein plays a role in the scission of the bud from the cell. It has been shown that M2 accumulates in the neck of the bud where scission occurs. Scission of the bud occurs independently of the host endosomal sorting complex required for transport (ESCRT) (35). Final release of the virus particles requires the enzymatic action of NA which cleaves the sialic acid moiety from the budding virus freeing it from the cell.
Viral pneumonia
One of the most serious complications of influenza A infections is the development of pneumonia. Viral induced pneumonia is a result of a combination of extensive viral replication in the lung, particularly the lower regions and the host immune response to the virus (36). The general progression to viral pneumonia development starts with viral replication in the lung epithelial cells and alveolar epithelium. Detection of viral infection in the epithelial cells induces the production and release of pro-inflammatory cytokines and chemokines (37). The host response to the release of these inflammatory mediators is the recruitment and activation of leukocytes from the circulatory system into the lung. In combination with resident alveolar macrophages the infiltration of mononuclear cells can lead to the overproduction of pro-inflammatory cytokines (38). Cell damage results from both the replicating virus which induces both necrosis and apoptosis in lung epithelial cells and macrophages and the products from activated leukocytes including reactive oxygen species (39). When the cell damage becomes extensive this leads to alveolar flooding where protein rich fluid accumulates in the airspace due to impaired fluid uptake from the alveolar epithelium (40). This edema is further exacerbated by an increased permeability of the lung capillaries in response to the infiltrating leukocytes. Damage to the blood air barrier results in decreased gas exchange which results in systemic hypoxia. Autopsy examination of lung tissue from influenza induced pneumonia shows signs of edema, hemorrhage, diffuse alveolar damage and the production of hyaline membrane (3). Acute respiratory distress syndrome (ARDS) leading to respiratory failure is the most common lethal development of influenza induced pneumonia. This syndrome is divided into three phases, the exudative, inflammatory and fibroproliferative phase (41). Damage to sufficient numbers of type II pneumocytes from influenza infection results in a decrease of surfactant production in the lung. Lung surfactant has been shown to have immunomodulating effects on immune cells (42). It has been demonstrated that during ARDS decreased lung surfactant results in increased PI3 kinase activity in immune cells (43). On lung epithelial cells and alveolar macrophages the decreased surfactant production results in increased AKT activation. For neutrophil dependent damage to the lung AKT activation has been shown to be involved (43). The ensuing alveolar flooding is a result of the impaired fluid removal across the lung epithelium due to impaired sodium transport across alveolar epithelial cells (41). In addition the increased cytokine and chemokine production of the alveolar epithelium increases the permeability of the lung endothelium allowing for increased fluid to enter the alveolar space (44). A further complicating factor with influenza induced pneumonia is the development of a secondary bacterial infection in the lungs (45). Up to half of pandemic influenza induced pneumonia was associated with a secondary bacterial infection (46). There is also an association with the type of bacteria found coinfecting with influenza A. Most commonly associated bacterial species which have been found to cause secondary infections to influenza are Streptococcus pneumoniae, Haemophilus influenza and Staphylococcus aureus (47). For these species of bacteria there is an apparent synergism with regards to virulence and pathogenicity. Coinfection with influenza or preinfection with influenza virus results in changes to the functioning of immune cells particularly neutrophils and alveolar macrophages (48). There is also an observed increase in bacterial adherence to the lung epithelium and a concurrent immunosuppression attributed to influenza infection (49). It has been observed in mice that influenza infection results in higher cytokine production of TNFα, IL-1β, IL-6, MIP1α and KC relative to a sole bacterial infection (50). Although influenza A infections predisposes the host to secondary bacterial pneumonia, a recent study has demonstrated that a previous infection with influenza affords some protection against subsequent bacterial pneumonia infection with a different influenza type. This protection was attributed to non-neutralizing antibody production from the initial infection against conserved internal viral protein NP (50).
Influenza A induced pro-inflammatory cellular signalling
Dendritic cells and macrophages, which reside in close proximity to the lung epithelium produce a significant amount of TNF-α and type I IFN in response to influenza virus infection (51). These cytokines activate intracellular signal transduction pathways in the lung epithelial cells. The highly pathogenic 1,918 Influenza virus induced gene expression profiles in mouse and non-human primate lungs which consisted of suppression of type 1 interferon antiviral response and enhanced expression of TNF-α, IFN-γ and IL-6 pro-inflammatory response. There is a dramatic increase in chemokines and inflammatory influx consisting of neutrophils, macrophages and T lymphocytes resulting in enhanced lung injury by a recombinant virus containing 1918 influenza genes HA and NA (52,53). Interferons activate intracellular Jak-Stat pathway as well as alternative signal transduction pathways in parallel (54,55), TNF-α activates a variety of protein kinases including c-Jun N-terminal kinase (JNK), extracellular-signal regulated kinases (ERK1/2), p38 and I-kappa B kinases (IKK). TNF-α activates nuclear factor kappaB (NF-κB) by means of a kinase relay module, involving the IκB kinase (IKK) α, β, γ signalosome (56). In addition to NF-κB, TNF-α also activates additional transcription factors such as AP1 (Fos-Jun) via JNK and activation transcription factor-2 (ATF-2) via JNK, ERK and p38 MAP kinase pathways (57-59). Involvement of ERK, JNK, p38 MAP kinases and activation of transcription factors AP1, NF-κB and ATF-2 has been described in response to influenza virus infection (60-64). Thus, multiple MAP kinase pathways are activated by cytokines as well as virus replication. Influenza virus propagation was impaired by inhibition of Raf/Mek/ERK pathway (60). Furthermore, lung-specific expression of active Raf kinase resulted in increased mortality of influenza virus infected mice (61). Targeting of NF-κB pathway for antiviral therapy in influenza infection has been shown to reduce viral titers and cytokine expression (65). Further understanding of intracellular signal transduction pathways may lead to the development of specific inhibitors of influenza viral replication as well as inflammatory host response that are involved in lung injury.
Intracellular detection of influenza A virus
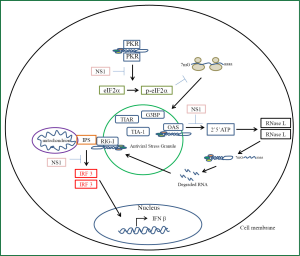
Cells of the lung epithelium have evolved receptors to detect the presence of viral nucleic acid and initiate a signaling cascade to induce an antiviral pro-inflammatory state (Figure 3). These receptors belong to a family of pattern recognition receptors each of which have a class of molecular structures which binds and activates them (66). Influenza A virus has been demonstrated to activate toll-like receptors (TLRs), NLRP3 inflammasome which belongs to the NOD-like receptors, RIG-I like receptors (RLR), PKR and 2'-5'-oligoadenylate synthetase (OAS) (67). Activation of these receptors is the primary signal for production of pro-inflammatory cytokines and chemokines which are released to recruit and activate leukocytes from the circulation to the site of influenza infection (68,69). The RLRs includes three members, retinoic acid inducible gene one (RIG-I), melanoma differentiation associated gene 5 (MDA5) and laboratory of general physiology 2 (LGP2). For cytosolic detection of influenza RIG-I is the primary receptor involved (70). The vRNP and replicative intermediates contain 5’triphosphates and double stranded RNA which are the unique viral molecular patterns that this receptor binds to distinguishing foreign viral RNA from self RNA (71). Activation of RIG-I results in a cascade of signaling events that pivots around the association with the mitochondrial outer membrane embedded protein IPS-1 (70). Subsequently the downstream signaling leads to the activation of transcription factors NFκB and interferon regulatory factors (IRF) 3 and 7. NFκB activation is responsible for the induction of transcription for pro-inflammatory cytokines while IRF3 and 7 induce the transcription of interferon beta gene which leads to the induction of antiviral genes to create an antiviral state (72,73). Similar to RIG-I signaling activation of TLR3 which is a receptor that resides in the endosomal membrane results in the activation of NFκB and the production of pro-inflammatory cytokines (74). TLR3 binds double stranded RNA. Unlike RIG-I in pulmonary epithelial cells this receptor does not contribute significantly to the production of interferon however its signaling significantly induces a pro-inflammatory state (75). In a mouse model of influenza induced pneumonia tlr3 negative mice were found to have a protective effect with reduced mortality compared to wild type mice with increased survival being attributed to a decreased production of pro-inflammatory cytokines (74). PKR is a 68 kDa latent protein expressed by most cells. It is induced by interferons and is activated by the binding of double stranded RNA or double stranded structures within ssRNA (76). PKR possess two double stranded RNA binding domains which binds its RNA ligand. Binding of RNA induces a conformational change that allows for monomeric PKR to form dimers which allows for autophosphorylation and activation of its kinase domain (77). Once activated PKR will phosphorylate its substrate eIF2α initiating a stress response in the cell (78). This stress response is a rapid reversible inhibition of protein translation. This reduction in translation leads to a reduced translation of all mRNA. The accumulation of mRNA with in the cytoplasm triggers the recruitment of three proteins; the cytotoxic granule associated RNA binding protein (TIA-1), TIA-1 cytotoxic granule associated RNA binding protein like 1 (TIAR) and GapSH3 domain binding protein (G3BP) (79). As these proteins bind the mRNA they trigger aggregation of these complexes together forming stress granules. PKR triggered inhibition of translation leads to the formation of these antiviral stress granules. Viral induced stress granules require PKR activation. Influenza induced stress granules recruit PKR, RIG-I, and oligoadenylate synthetase (OAS) (80). This platform provides an efficient mechanism for interferon induction in response to viral infection. It has also been shown that vRNA colocalizes with these granules which activates RIG-I. Once activated the antiviral stress granule is recruited to the mitochondria to associate with IPS-1. The viral NS1 protein has been shown to antagonize interferon induction at many levels. This viral protein inhibits activation of PKR, OAS, and RIG-I (81). Through inhibition of PKR activation influenza A virus prevents antiviral stress granule formation and production of interferons while the proinflammatory arm of signaling remains intact. The OAS/RNAse L system is part of the innate immune response. OAS is a pattern recognition receptor that is activated by viral double stranded RNA (82,83). Activation of OAS leads to the production of a unique molecule 2',5' oligoadenylate. This molecule specifically activates RNAse L (84). Typically OAS produces trimers and tetramers of 2',5'-oligoadenlyate. Activation of RNase L requires the formation of dimers in response to binding of 2',5'-oligoadenlyate. The active dimer cleaves single stranded RNA at the dinucleotide motif of uracil adenosine or uracil uracil (85). The resulting cleavage product possesses a 3' phosphate which acts as a substrate for RIG-I thus amplifying the signaling to interferon production (67,86). In addition the degradation of viral RNA coupled with the inhibition of translation is the main mechanism of RNase L antiviral activity. However RNase L is not specific to viral RNA degradation and it degrades host rRNA and mRNA and can lead to apoptosis. The NS1 protein of Influenza A has evolved to inhibit the action of the OAS/RNase L system (81). NS1 has been shown to sequester viral double stranded RNA so that it prevents activation of OAS and as a result its effector protein RNase L (87).
Conclusions
Since the influenza A virus is comprised of multiple strains that continually mutate, further study is required in the identification of the host factors that are absolutely necessary for viral replication. This area of research is showing progress in identifying new therapeutic targets to act as adjuncts to the seasonal vaccines and these targets may prove to be less susceptible to viral mutations.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Centers for Disease Control and Prevention (CDC). Estimates of deaths associated with seasonal influenza --- United States, 1976-2007. MMWR Morb Mortal Wkly Rep 2010;59:1057-62. [PubMed]
- Dawood FS, Iuliano AD, Reed C, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 2012;12:687-95. [PubMed]
- Howard WA, Peiris M, Hayden FG. Report of the ‘mechanisms of lung injury and immunomodulator interventions in influenza’ workshop, 21 March 2010, Ventura, California, USA. Influenza Other Respi Viruses 2011;5:453-4, e458-75.
- Belser JA, Maines TR, Tumpey TM, et al. Influenza A virus transmission: contributing factors and clinical implications. Expert Rev Mol Med 2010;12:e39. [PubMed]
- Yang W, Elankumaran S, Marr LC. Concentrations and size distributions of airborne influenza A viruses measured indoors at a health centre, a day-care centre and on aeroplanes. J R Soc Interface 2011;8:1176-84. [PubMed]
- Lakdawala SS, Subbarao K. The ongoing battle against influenza: The challenge of flu transmission. Nat Med 2012;18:1468-70. [PubMed]
- Yang W, Marr LC. Dynamics of airborne influenza A viruses indoors and dependence on humidity. PLoS One 2011;6:e21481. [PubMed]
- Yang W, Elankumaran S, Marr LC. Relationship between humidity and influenza A viability in droplets and implications for influenza’s seasonality. PLoS One 2012;7:e46789. [PubMed]
- Azziz Baumgartner E, Dao CN, Nasreen S, et al. Seasonality, timing, and climate drivers of influenza activity worldwide. J Infect Dis 2012;206:838-46. [PubMed]
- Eccles R. Understanding the symptoms of the common cold and influenza. Lancet Infect Dis 2005;5:718-25. [PubMed]
- Van Reeth K. Cytokines in the pathogenesis of influenza. Vet Microbiol 2000;74:109-16. [PubMed]
- Coloma R, Valpuesta JM, Arranz R, et al. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog 2009;5:e1000491. [PubMed]
- Fournier E, Moules V, Essere B, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res 2012;40:2197-209. [PubMed]
- Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J 1997;16:1248-57. [PubMed]
- Zheng W, Tao YJ. Structure and assembly of the influenza A virus ribonucleoprotein complex. FEBS Lett 2013;587:1206-14. [PubMed]
- Gerl MJ, Sampaio JL, Urban S, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol 2012;196:213-21. [PubMed]
- Schaap IA, Eghiaian F, des Georges A, et al. Effect of envelope proteins on the mechanical properties of influenza virus. J Biol Chem 2012;287:41078-88. [PubMed]
- Bertram S, Glowacka I, Steffen I, et al. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol 2010;20:298-310. [PubMed]
- Hamilton BS, Gludish DW, Whittaker GR. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J Virol 2012;86:10579-86. [PubMed]
- Upham JP, Pickett D, Irimura T, et al. Macrophage receptors for influenza A virus: role of the macrophage galactose-type lectin and mannose receptor in viral entry. J Virol 2010;84:3730-7. [PubMed]
- Leung HS, Li OT, Chan RW, et al. Entry of influenza A Virus with a α2,6-linked sialic acid binding preference requires host fibronectin. J Virol 2012;86:10704-13. [PubMed]
- Fontana J, Cardone G, Heymann JB, et al. Structural changes in Influenza virus at low pH characterized by cryo-electron tomography. J Virol 2012;86:2919-29. [PubMed]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008;451:591-5. [PubMed]
- Lee KK. Architecture of a nascent viral fusion pore. EMBO J 2010;29:1299-311. [PubMed]
- Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol 1991;65:232-44. [PubMed]
- O’Neill RE, Jaskunas R, Blobel G, et al. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem 1995;270:22701-4. [PubMed]
- Umbach JL, Yen HL, Poon LL, et al. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. MBio 2010;1:e00204-10. [PubMed]
- Perez JT, Zlatev I, Aggarwal S, et al. A small-RNA enhancer of viral polymerase activity. J Virol 2012;86:13475-85. [PubMed]
- Widjaja I, de Vries E, Rottier PJ, et al. Competition between influenza A virus genome segments. PLoS One 2012;7:e47529. [PubMed]
- Yu M, Liu X, Cao S, et al. Identification and characterization of three novel nuclear export signals in the influenza A virus nucleoprotein. J Virol 2012;86:4970-80. [PubMed]
- Cao S, Liu X, Yu M, et al. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. J Virol 2012;86:4883-91. [PubMed]
- Kawaguchi A, Matsumoto K, Nagata K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J Virol 2012;86:11086-95. [PubMed]
- Wang S, Li H, Chen Y, et al. Transport of influenza virus neuraminidase (NA) to host cell surface is regulated by ARHGAP21 and Cdc42 proteins. J Biol Chem 2012;287:9804-16. [PubMed]
- Kerviel A, Thomas A, Chaloin L, et al. Virus assembly and plasma membrane domains: which came first? Virus Res 2013;171:332-40. [PubMed]
- Rossman JS, Jing X, Leser GP, et al. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010;142:902-13. [PubMed]
- Ruuskanen O, Lahti E, Jennings LC, et al. Viral pneumonia. Lancet 2011;377:1264-75. [PubMed]
- Herold S, von Wulffen W, Steinmueller M, et al. Alveolar epithelial cells direct monocyte transepithelial migration upon influenza virus infection: impact of chemokines and adhesion molecules. J Immunol 2006;177:1817-24. [PubMed]
- Geiler J, Michaelis M, Sithisarn P, et al. Comparison of pro-inflammatory cytokine expression and cellular signal transduction in human macrophages infected with different influenza A viruses. Med Microbiol Immunol 2011;200:53-60. [PubMed]
- Carnesecchi S, Pache JC, Barazzone-Argiroffo C. NOX enzymes: potential target for the treatment of acute lung injury. Cell Mol Life Sci 2012;69:2373-85. [PubMed]
- Wolk KE, Lazarowski ER, Traylor ZP, et al. Influenza A virus inhibits alveolar fluid clearance in BALB/c mice. Am J Respir Crit Care Med 2008;178:969-76. [PubMed]
- MacCallum NS, Evans TW. Epidemiology of acute lung injury. Curr Opin Crit Care 2005;11:43-9. [PubMed]
- Hawgood S, Brown C, Edmondson J, et al. Pulmonary collectins modulate strain-specific influenza a virus infection and host responses. J Virol 2004;78:8565-72. [PubMed]
- Mittal N, Sanyal SN. Effect of exogenous surfactant on phosphatidylinositol 3-kinase-Akt pathway and peroxisome proliferator activated receptor-γ during endotoxin induced acute respiratory distress syndrome. Mol Cell Biochem 2012;361:135-41. [PubMed]
- Armstrong SM, Wang C, Wang Y, et al. Influenza Primes Human Lung Microvascular Endothelium To Leak Upon Exposure To Staphylococcus Aureus. In: Anonymous. eds. American Thoracic Society International Conference Abstracts 2013;187:Meeting Abstracts.
- Metersky ML, Masterton RG, Lode H, et al. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis 2012;16:e321-31. [PubMed]
- Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008;198:962-70. [PubMed]
- Liderot K, Ahl M, Ozenci V. Secondary Bacterial Infections in Patients with Seasonal Influenza A and Pandemic H1N1. Biomed Res Int 2013;2013:376219.
- Giamarellos-Bourboulis EJ, Raftogiannis M, Antonopoulou A, et al. Effect of the novel influenza A (H1N1) virus in the human immune system. PLoS One 2009;4:e8393. [PubMed]
- van der Sluijs KF, van Elden LJ, Nijhuis M, et al. IL-10 is an important mediator of the enhanced susceptibility to pneumococcal pneumonia after influenza infection. J Immunol 2004;172:7603-9. [PubMed]
- Haynes L, Szaba FM, Eaton SM, et al. Immunity to the conserved influenza nucleoprotein reduces susceptibility to secondary bacterial infections. J Immunol 2012;189:4921-9. [PubMed]
- Julkunen I, Sareneva T, Pirhonen J, et al. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev 2001;12:171-80. [PubMed]
- Kash JC, Tumpey TM, Proll SC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 2006;443:578-81. [PubMed]
- Kobasa D, Jones SM, Shinya K, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007;445:319-23. [PubMed]
- Ramana CV, Gil MP, Schreiber RD, et al. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol 2002;23:96-101. [PubMed]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 2005;5:375-86. [PubMed]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell 2002;109:S81-96. [PubMed]
- Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol 2001;11:372-7. [PubMed]
- Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 2001;104:487-501. [PubMed]
- Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 2004;6:97-105. [PubMed]
- Pleschka S, Wolff T, Ehrhardt C, et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol 2001;3:301-5. [PubMed]
- Olschläger V, Pleschka S, Fischer T, et al. Lung-specific expression of active Raf kinase results in increased mortality of influenza A virus-infected mice. Oncogene 2004;23:6639-46. [PubMed]
- Conze D, Lumsden J, Enslen H, et al. Activation of p38 MAP kinase in T cells facilitates the immune response to the influenza virus. Mol Immunol 2000;37:503-13. [PubMed]
- Ludwig S, Ehrhardt C, Neumeier ER, et al. Influenza virus-induced AP-1-dependent gene expression requires activation of the JNK signaling pathway. J Biol Chem 2001;276:10990-8.
- Flory E, Kunz M, Scheller C, et al. Influenza virus-induced NF-kappaB-dependent gene expression is mediated by overexpression of viral proteins and involves oxidative radicals and activation of IkappaB kinase. J Biol Chem 2000;275:8307-14. [PubMed]
- Pinto R, Herold S, Cakarova L, et al. Inhibition of influenza virus-induced NF-kappaB and Raf/MEK/ERK activation can reduce both virus titers and cytokine expression simultaneously in vitro and in vivo. Antiviral Res 2011;92:45-56. [PubMed]
- Bleiblo F, Michael P, Brabant D, et al. The role of immunostimulatory nucleic acids in septic shock. Int J Clin Exp Med 2012;5:1-23. [PubMed]
- Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 2012;86:2900-10. [PubMed]
- Mahalingam S, Karupiah G. Chemokines and chemokine receptors in infectious diseases. Immunol Cell Biol 1999;77:469-75. [PubMed]
- Zhou J, Law HK, Cheung CY, et al. Differential expression of chemokines and their receptors in adult and neonatal macrophages infected with human or avian influenza viruses. J Infect Dis 2006;194:61-70. [PubMed]
- Loo YM, Fornek J, Crochet N, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 2008;82:335-45. [PubMed]
- Pichlmair A, Schulz O, Tan CP, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 2006;314:997-1001. [PubMed]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 2006;6:644-58. [PubMed]
- Pauli EK, Schmolke M, Wolff T, et al. Influenza A virus inhibits type I IFN signaling via NF-kappaB-dependent induction of SOCS-3 expression. PLoS Pathog 2008;4:e1000196. [PubMed]
- Le Goffic R, Balloy V, Lagranderie M, et al. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog 2006;2:e53. [PubMed]
- Le Goffic R, Pothlichet J, Vitour D, et al. Cutting Edge: Influenza A virus activates TLR3 dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol 2007;178:3368-72. [PubMed]
- Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA 2004;10:1934-45. [PubMed]
- Wu S, Kaufman RJ. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J Biol Chem 1997;272:1291-6. [PubMed]
- Samuel CE. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem 1993;268:7603-6. [PubMed]
- Khaperskyy DA, Hatchette TF, McCormick C. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J 2012;26:1629-39. [PubMed]
- Onomoto K, Jogi M, Yoo JS, et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 2012;7:e43031. [PubMed]
- Österlund P, Strengell M, Sarin LP, et al. Incoming influenza A virus evades early host recognition, while influenza B virus induces interferon expression directly upon entry. J Virol 2012;86:11183-93. [PubMed]
- Silverman RH. Viral encounters with 2’,5’-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 2007;81:12720-9. [PubMed]
- Anderson BR, Muramatsu H, Jha BK, et al. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res 2011;39:9329-38. [PubMed]
- Zhou A, Hassel BA, Silverman RH. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 1993;72:753-65. [PubMed]
- Wreschner DH, McCauley JW, Skehel JJ, et al. Interferon action--sequence specificity of the ppp(A2'p)nA-dependent ribonuclease. Nature 1981;289:414-7. [PubMed]
- Malathi K, Dong B, Gale M Jr, et al. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007;448:816-9. [PubMed]
- Hale BG, Randall RE, Ortín J, et al. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 2008;89:2359-76. [PubMed]


