CRISPR-barcoding in non small cell lung cancer: from intratumor genetic heterogeneity modeling to cancer therapy application
Cancers are composed of different populations of cells with distinct molecular and phenotypic characteristics that could have potential impact on cancer progression, treatment and risk of disease relapse (1). Genomic studies have demonstrated that human cancers comprise a molecular mosaic of cells, spatially and temporally different (2).
The intratumor genetic heterogeneity represents one of the most clinical challenges associated to the drug resistance. The acquired resistence may occur through different mechanism, including the acquisition of de novo mutations during cancer therapy and the rare resistant clones pre-exsist in the tumor before the treatment (3). The detection of the pre-existing resistant subclones is really hard since they are very rare, therefore the sensitivity of the current approaches is generally insufficient to comprehensively assess cancer individual cells in heterogeneous cancer-cell populations (4).
Clustered regularly interspaced short palindromic repeats (CRISPR) associated protein 9 (Cas9) system is a DNA editing technology that has revolutionized the field of genetic engineering and may be useful to identify different cells with specific molecular features.
CRISPR-Cas9 system is a DNA editing tool based on an RNA guided DNA endonuclease that requires a short guide RNA (sgRNA) to recognize specific target genomic sequence (5). In details, the nuclease Cas9 is guided by sgRNA that hybridizes to the complementary targeted nucleotides, thus Cas9 cleaves the genomic sequence of interest producing DNA double-strand breaks (DSBs), triggering the cellular mechanisms of DNA repair, including non-homologous end joining (NHEJ) or homologous directed repair (HDR). NHEJ frequently results in the insertion or deletion of a few nucleotides and it can be used to knockout the gene of interest thought frameshift mutations. Repair by HDR include a donor DNA template displaying sequence homology to the targeted locus and it can be exploited to modify a gene by introducing point mutations or to insert more extensive modifications achieve of the genome (6).
CRISPR-Cas9 system showed higher specificity and efficiency compared to other DNA editing technologies based on protein-DNA recognition, including zinc finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN) (7).
Furthermore, further innovations of CRISPR/Cas9 technology allowed to expand its application in several fields including cancer research.
Recently, Guernet et al. developed high complex CRISPR-barcoding system as an alternative tool to the classical lentiviral DNA barcode libraries, ensuring the detection of thousands of distinct barcodes through qPCR or deep-sequencing. This new technology enables a high-resolution tracking of single specific cancer cells allowing to identify even rare pre-existing resistant subclones potentially involved in mechanisms of acquired resistance to therapy (6).
Guernet et al. highlighted particularly the use of CRISPR-barcoding to develop models of drug resistance in non-small cell lung cancer (NSCLC) since this system closely mimics the clonal dynamics of cancer. They generated models of NSCLC resistance to EGFR inhibitors based on a specific sgRNA and a donor single-stranded DNA oligonucleotide (ssODN) containing as barcodes different genetic aberrations, including the EGFR T790M mutation, a secondary mutation in the catalytic domain associated to acquired resistance and KRAS G12D mutation, a well-known negative predictor for primary responsiveness to EGFR inhibitors. Studies in immunocompromised mice injected with KRAS-G12D and EGFR-T790M CRISPR-barcoded cells showed an increase of both mutations in the tumors from gefitinib treated mice, demonstrating that resistant subclones were selected (6).
Similarly, they proposed another model based on CRISPR-barcoding specific for the rearrangement EML4-ALK suggested that this aberration could represent a novel mechanism of resistance to EGFR inhibitors.
The discovery of the druggable protein kinases, and the specific tyrosine kinase inhibitor (TKI) has greatly revolutionized the treatment of NSCLC, offering a substantial improvement of outcomes compared with standard chemotherapy. NSCLC, patients harboring EGFR L858R point mutation and exon 19 deletions are sensitive to generation EGFR TKIs such as gefitinib, erlotinib and afatinib (8). Similarly, patients carrying ALK rearrangements define a unique molecular subset of NSCLC responsive to ALK TKI crizotinib (8).
Both EGFR and ALK TKIs have been approved as standards of care for NSCLC patients with these genetic aberrations. Unfortunately, despite the initial benefit, the long-term effectiveness of target therapies is limited since the patients develop drug resistance through a variety of mechanisms.
The clinical experience in the treatment with first- and second-generation EGFR and ALK TKIs in NSCLC patients suggested that the overcoming of the TKIs resistance is the main challenge in this clinical setting. To date, several different TKI resistance mechanisms have been identified within EGFR-mutant and ALK-rearranged patients, including secondary mutations in the kinase target, gene amplification of the primary oncogene, and upregulation of bypass signaling tracts (9,10).
Previous studies showed that different mechanisms of resistance can coexist within the same tumor or in metastases from the same patients before the onset of therapy. The response to treatment could be conditioned from integrating mechanisms used by cancer cell clones to escape therapy, therefore TKIs resistance represents a dynamic and multifactorial process.
NSCLC could composed of multiple subclones that can be selected and drive disease progression, especially under selection pressures, such as the treatment with specific TKIs.
In this context, CRISPR-barcoding could represent a useful assay to study intratumor molecular heterogeneity in NSCLC model cancer in order to identify different subclonal genetic aberration that could lead to an heterogeneous response and the TKIs resistance.
Consistent with previous data obtained by Guernet et al., further multiplex modeling based on CRISPR-barcoding could be set up to analyze the clonal dynamics in NSCLC and the resistance to TKI in order to evaluate the efficacy of combined drug and the optimal therapeutic setting in NSCLC (Figure 1).
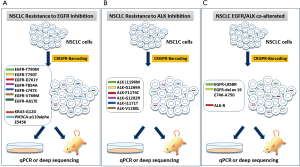
Multiplex model could reproduces the potential crosstalk between distinct cancer cells, even among rare pre-existant subclones within a tumor mass, thus providing a model that could recapitulates the complexity of the heterogeneous response to TKIs.
Furthermore, CRISPR-barcoding could represent an optimal system to clarify the responsiveness and resistance to specific TKIs in NSCLC patients that harboring concomitant EGFR/ALK alterations.
EGFR-mutations and ALK-rearrangement are generally mutually exclusive, however these aberrations can coexist in a small subgroup of NSCLC patients that have diverse responses to specific TKIs, however few contrasting data have been currently reported (11-14)
Model based on CRISPR-barcoding could help to understand different mutation tumor burden underlying heterogeneous responsiveness to TKIs in EGFR/ALK co-alterated patients (Figure 1C).
In conclusion, CRISPR-barcoding could have interesting clinical implications in cancer therapy application, especially to overcome the intratumor molecular heterogeneity since it allows the detection of rare pre-existing clones that might plant the seeds for drug resistence.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational Implications of Tumor Heterogeneity. Clin Cancer Res 2015;21:1258-66. [Crossref] [PubMed]
- Bria E, Pilotto S, Amato E, et al. Molecular heterogeneity assessment by next-generation sequencing and response to gefitinib of EGFR mutant advanced lung adenocarcinoma. Oncotarget 2015;6:12783-95. [Crossref] [PubMed]
- Turke AB, Zejnullahu K, Wu YL, et al. Pre-existence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. [Crossref] [PubMed]
- Bhang HE, Ruddy DA, Krishnamurthy Radhakrishna V, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med 2015;21:440-8. [Crossref] [PubMed]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods 2013;10:957-63. [Crossref] [PubMed]
- Guernet A, Mungamuri SK, Cartier D, et al. CRISPR-barcoding for intratumor genetic heterogeneity modeling and functional analysis of oncogenic driver mutations. Mol Cell 2016;63:526-38. [Crossref] [PubMed]
- Chen S, Sun H, Miao K, et al. CRISPR-Cas9: from Genome Editing to Cancer Research. Int J Biol Sci 2016;12:1427-36. [Crossref] [PubMed]
- Maione P, Sacco PC, Sgambato A, et al. Overcoming resistance to targeted therapies in NSCLC: current approaches and clinical application. Ther Adv Med Oncol 2015;7:263-73. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J Clin Oncol 2013;31:3987-96. [Crossref] [PubMed]
- Liao BC, Lin CC, Shih JY, et al. Treating patients with ALK-positive nonsmall cell lung cancer: latest evidence and management strategy. Ther Adv Med Oncol 2015;7:274-90. [Crossref] [PubMed]
- Zito Marino F, Morabito A, Gridelli C, et al. Crizotinib Response in a Late Relapse of ALK-positive Lung Adenocarcinoma. Appl Immunohistochem Mol Morphol 2016;24:e86-e88. [Crossref] [PubMed]
- Kuo YW, Wu SG, Ho CC, et al. Good response to gefitinib in lung adenocarcinoma harboring coexisting EML4-ALK fusion gene and EGFR mutation. J Thorac Oncol 2010;5:2039-40. [Crossref] [PubMed]
- Popat S, Gonzalez D, Min T, et al. ALK translocation is associated with ALK immunoreactivity and extensive signet-ring morphology in primary lung adenocarcinoma. Lung Cancer 2012;75:300-5. [Crossref] [PubMed]
- Tiseo M, Gelsomino F, Boggiani D, et al. EGFR and EML4-ALK gene mutations in NSCLC: a case report of erlotinib-resistant patient with both concomitant mutations. Lung Cancer 2011;71:241-3. [Crossref] [PubMed]


