Epigenetic aberrant methylation of tumor suppressor genes in small cell lung cancer
Introduction
Lung cancer continues to be a huge burden on the health status of Chinese and people worldwide, representing 15% of the new cases of all cancers diagnosed in 2008 (1). Lung cancer includes a group of heterogeneous histological types in terms of clinicopathological features and is commonly classified into two major subgroups, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). SCLC comprises nearly 20% of lung cancer with different biological behavior, clinical characteristics, responsiveness to chemotherapy and prognosis of patients (2). The malignant cells exhibit scarce cytoplasm, molded nuclei and neuroendocrine (NE) differentiation, as evidenced by expressing a variety of NE substance. But SCLC is also different from carcinoid in aspects of carcinogenesis and pathogenesis. Although the radio-chemotherapy and surgical resection for SCLC have considerable improvement, the patients still have poor prognosis with cumulative survival rate of around 5% at 5 years (3). The latest advances in molecular biological technologies have found some mechanisms of malignant neoplasia. In brief, SCLC cells show the abilities of continuos proliferation, evasion of apoptosis, local invasion and remote metastasis, which is related to silence of tumor suppressor genes (TSGs). Genetic and epigenetic alteration both are responsible for inactivation of TSGs. Epigenetic aberration, one of the earliest and most common phenomena in human malignancies, could lead to abrogation of various TSGs. Studies about epigenetic alteration could be contributed to increasing the rate of early stage at diagnosis, and therefore, the patients with SCLC could benefit from higher early detection possibilities with good responsiveness to intervention and long-time survival.
Expression of TSGs is regulated by the gene promotors, while CpG islands are commonly seen in the up-stream of gene promotor. The clusters of guanine and cytosine (GC) are ranged from 0.5 to 5 kbp, with high ratio of GC (60-70%) (4). Almost fifty percent of the genes have the so-called CpG islands in the entire genome. About eighty percent of the CpG islands are hyper-methylated and related to repetitive elements in the human DNA. The novel thing is that such CpG islands located at the region of gene promoters are usually un-methylated in normal cells (5). Methylation of CpG islands at their cytosine 5 position is characterized by gene, tissue and differentiation specificities. Nevertheless, the mechanisms of gene silence via promoter methylation in transcription level are not fully understood. Current literatures have reported involved mechanisms as follow: (I) DNA methylation may impede expression of house keep genes necessary for epigenetic reprogramming (6); (II) Methylated DNA are interacted with some regulatory proteins, including methylated DNA binding proteins (MBPs), MBPs like methyl-CpG binding protein 2 (MeCP2), and histone deacetylases (HDACs) (7); (III) Histone methylation via DNA methyltransferases (DNMTs) have additional chromatin modifications and mediate transcriptional repression (8).
Individual tumor types have their characteristic pattern of acquired aberrant methylation. Although many genes inactivated by methylation have been found in NSCLC, differences in molecular and genetic pathogenesis between SCLC and NSCLC can not be ignored. Data about aberrant methylation of TSGs in SCLC are limited. Aberrant hyper-methylated genes could be prognostic factors for SCLC, which need to be identified. In the present review article, we summarize the epigenetic methylation of key TSGs in SCLC. In addition, we will also provide a brief overview of the potential of these alterations as tumor markers and therapeutic targets for SCLC.
TSGs silenced by methylation in SCLC
Cell cycle control genes
P16INK and P14ARF, located on chromosome 9p21, both are transcripts of the cyclin-dependent kinase inhibitor 2A (CDKN2A), which are silenced via methylated in multiple tumors (9-11). Mitsuo Sato et al. (12) had reported comprehensive hyper-methylation profile of the p14ARF and p16INK4a genes in 12 SCLC cell lines. In SCLC, P16INK was methylated in about 31-37% of primary or metastatic tumors tissues (11,13). The hyper-methylated promoter was found at the frequency of 20% in all promoters for primary tumors samples and 13% for metastatic tumor samples. In SCLC cell lines, methylated P16INK gene was detected in all promoter in H1618 cells. P16INK gene was un-methylated in H82 and H146 cells (11). Whereas, P14ARF was methylated at a low frequency of 6% (12). Repression of p14ARF is mainly due to homozygous deletion or mutation instead of heterozygosis methylation. Even Mitsuo Sato et al. (12) found there was no methylated promoter of p14ARF gene in 12 kinds of SCLC cell lines. Those results indicated that hyper-methylation of p14ARF promoter is rare in SCLC. But promoter methylation is the predominant mechanism for P16INK inactivation.
The RAS association domain family 1A (RASSF1A) (also known as NORE2A, 123F2, RDA32 and REH3P21) is a gatekeeper for G1/S cell cycle progression, which regulates diverse cellular functions (14). Silence of RASSF1A leads to immortal cells proliferation and impeding apoptosis, which can serve as a useful diagnostic marker of cancers. M. SATO et al. (13) had detected the methylation status of RASSF1A gene in SCLC cells. The bisulfite genomic sequencing analysis showed RASSF1A promoters were complete methylated in 10 of 12 SCLC cell lines and incomplete methylated in 1 of 12 SCLC cell lines. RASSF1A is homogeneous deletion only in one SCLC cell line: H740 cells. Although Loss of heterozygosity (LOH) at 3p21.3 occurs in more than 90% of SCLCs, Dammann R et al. (15) had demonstrated the CpG islands around RASSF1A promoters, located at 3p21.3, were hyper-methylated in 22 of 28 primary SCLC tissues. Pelosi G et al. (16) also found the expression level of RASSF1A mRNA is closely correlated with methylation degree of promotors. Further analysis indicated RASSF1A hyper-methylation enabled a highly sensibility and relative specificity between patients with and without SCLC (17). Methylated RASSF1A gene detection may be a promising molecular tool for diagnosis of primary SCLC.
Apoptotic genes
Death associated protein kinase (DAPK) gene is a novel tumor suppressor gene, which transmits apoptotic signals and malignant transformation (18). DAPK induces cell apoptosis through extensive intracellular signaling pathways, including serine/threonine kinase signal pathway, tumor necrosis factor-α (TNF-α) signal pathway, tumor growth factor-β (TGF-β) signal pathway, Fas ligand signal pathway and caspase signal pathway. However, apoptosis is only one side of duality of cell death. Autophagy, type II cell death (type I being the apoptotic cell death), was long ignored by the majority of the scientists. It exists in all organisms, especially under the malnutrition condition. Expression of DAPK, a potent cell death protein, also triggers the type II cell death process with autophagic characteristics such as autophagic vesicles and autolysosomes in cytoplasm. In most of human tumors, down-regulated expression of DAPK was not due to DNA deletion, rearrangement of DNA or LOH, but due to the epigenetic silencing by aberrant methylation. Promoter methylation of DAPK was detected by Esteller M and his colleagues in lung cancer (11). The frequency of methylation is 16%. The loss of DAPK expression acts as a molecular switch involved in disfunction of cell death like apoptosis and autophagy, which leads to pathogenesis and carcinogenesis.
Apoptosis-associated speck-like protein containing a CA spase recruitment domains (ASC) gene, protein kinase C delta binding protein (PRKCDBP) gene and Wilms tumor 1 (WT1) gene both are involved in cell apoptosis via various cellular signal pathways. The expression of those genes in SCLC was blocked compared to normal lung (19). Fukasawa M et al. (19) found those gene both were hyper-methylated in SCLC cell lines using the promoter associated methylated DNA amplification and DNA chip (PMAD) method. Hyper-methylation of the ASC gene promotor was detected in LK79cells, S2cells and SBC-3 cells, PRKCDBP gene in S2cells and SBC-3 cells and WT1 gene in LK79cells and SBC-3 cells. The average hyper-methylation rate was 14.0% for SCLC (LK79, S-2, SBC-3 cells). Target of methylation induced silencing (TMS1) gene, also known as ASC gene, encodes a 22-kDa CA spase recruitment domains (CARD) protein, which acts as a adapter protein. This adapter protein actives caspases through cleavage of proteins pathway via CARD domains. Activated caspases stimulate a family of cysteine proteases and trigger programmed cell death. However, the ability of response to caspases can be lost due to epigenetic silence of TMS1 gene in SCLC cells (20). Methylation-specific polymerase chain reaction (MSP) test indicated that aberrant methylation of TMS1 was present in 70% (40 of 57) of SCLC cell lines and 41% (13 of 32) of SCLC tumor tissues (20).
Caspase-8 is a key component of apoptotic complexes. Fulda S et al. (21) reported the caspase-8 mRNA expression was in different levels in SCLCs. The expression levels of caspase-8 protein were associated with the methylation status of a CpG-rich part of the 5' flanking region of caspase-8. In VH-64 cells with a relative high level of caspase-8, only the unmethylated CpG islands of caspase-8 promotor were found. While in cell lines with undetectable levels of caspase-8 expression like CADO cells, CpG islands of caspase-8 were detected only in the methylated form.
DNA repair genes
DNA repair plays a critical role in the regulation of various physiological or pathological conditions and remove of mutagenic and cytotoxic production. O6-methylguanine DNA methyltransferase (MGMT) gene, encoding O6-AG DNA alkyltransferase, has also been implied to mediate DNA repair by remove of alkyl groups from the O6 position of guanine. Few scholars had reported epigenetic silencing of MGMT gene via hyper-methylation of promoter leads to loss of MGMT activity (11,22-24). Methylation of MGMT promoter was found in 36 of 39 (92%) SCLC cell lines (23). The frequency of methylated MGMT genes in SCLC tissues was 16% (24). However, studies of Esteller M (25) had opposite results. Promoter hyper-methylation of MGMT gene was a common event in NSCLC samples (29%). There was no epigenetic methylation in SCLC samples.
Fragile histidine triad (FHIT) gene is a DNA repair gene, which loss of expression is frequent in SCLC. In addition, the correlation between FHIT methylation and lack of FHIT expression was highly significant (P<0.0001) (26). The frequency of methylated FHIT gene is 64% in SCLC samples (24).
Metastasis ralated genes
Cadherin 1 (CDH1), cadherin 13 (CDH13), deleted in liver cancer 1 (DLC1) and tissue inhibitor of metalloproteinase 3 (TIMP3) both are classic TSGs and key cell adhesion molecules to maintain normal tissue architecture and inhibit tumor initiation, adhesion and metastasis. CDH1 gene was methylated in 17 of 39 (44%) SCLC cell lines (23), while methylation of CDH13 promoter was at a higher frequency of 92% (36 of 39). And other reports had different results. Zöchbauer-Müller S et al. reported the frequency of methylated CDH13 gene was only 15% (24). Fukasawa M et al. (19) found CDH13 was methylated in LK-79 and SBC-3 cells, but not in S2 cells. And their studies also indicted that methylated DLC1 gene was only found in SBC-3 cells. Esteller M and his colleagues (11) found the frequency of hyper-methylated TIMP-3 gene was 19% in lung cancer tissues.
Other TSGs
Similarly, other TSGs related to carcinogenesis silenced by promoter methylation, such as adenomatous polyposis coli (APC) (23,24,26), zinc finger (Blu) (27), calcitonin-related polypeptide alpha (CALCA) (19), epidermal growth factor like domain 7 (EGFL7) (15), endothelin B receptor (ETBR) (28), fatty acid binding protein 3 (FABP3) (19), GATA binding protein 4 (GATA4) (29), GATA binding protein 5 (GATA5) (29), glutathione S transferase pi 1 (GSTP1) (11,23,24), hyper-methylated in cancer 1 (HIC1) (19), 5-hydroxytryptamine receptor 1B (HTR1B) (19), interferon regulatory factor 7 (IRF7) (19), low density lipoprotein-related protein 2 (LRP2) (19), myosin 18B (MYO18B) (30), myogenic differentiation 1 (MYOD1) (19), neuron-restrictive silencer factor (NRSF) (31), paired box gene 3 (PAX3) (19), receptor interacting serine threonine kinase 3 (RIPK3) (19), retinoic acid receptor beta (RAR-β) (19,23,24,32), Ras-related associated with diabetes (RRAD) (33), sex determining region Y box 18 (SOX18) (15) and thyroid hormone receptor β1 (TRβ1) (34) have already been identified in SCLC (Table 1).
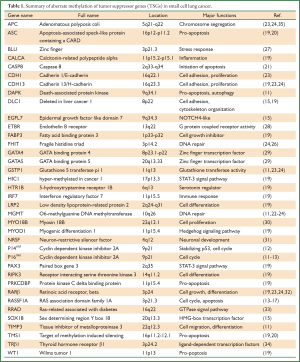
Full Table
Above results may cause considerable controversies about the degree and frequency of TSGs methylation in SCLC. The frequency of some TSGs methylation varies widely in different literatures. These studies reflect differences between primary SCLC tissues and cell lines. The degree of methylation also differ among cell lines with differentiation subtypes. Consistent with this, the compatibility, veracity and reliability of detected methods need to be demonstrated.
Clinical application of methylated TSGs
Analysis of methylation of TSGs has a huge application foreground with unparalleled advantages. First, DNA is easier to extract from blood, biological fluid, tumor mass and formaldehyde-fixed samples due to stability of DNA. Second, methylation-specific PCR has relatively high sensitivity and a DNA-based methylated marker can be detected among one thousand unmethylated alleles (36). Third, methylated DNA test is a noninvasive diagnostic method. Fourth, methylation of TSGs occurs in early stage of SCLC, which can be used for screening. Furthermore, methylation of TSGs can be reserved by methylation reagent, like 5-Aza-2'-deoxycytidine and its analogues (21,35) and TSGs methylation is correlated with response to chemotherapy (22).
According to the literature (17), methylated RASSF1A promoters were detected in 35 of 40 bronchial aspirates samples collected from patients with SCLC. However, RASSF1A hyper-methylation was not founded in 0 of 46 samples from patients with non-neoplastic lung disease. Moreover, the frequency of RASSF1A methylation increases along with poor differentiation of SCLC cells. Methylation of TSGs can be prognostic predictor for patient with SCLC. The 2-year survival for patients who were positive and negative for RAR-β methylation was significantly different in SCLC patients (chi-square test, P=0.044) (32). Reactivation of TSGs by 5-Aza-2'-deoxycytidine might be an effective strategy to intervene in SCLC (21,35). Pietanza MC et al. (22) found patients with methylated MGMT had higher complete response rate or partial response rate compared to those with unmethylated MGMT (38% vs. 7%) to temozolomide. Restoration of sensitivity for chemotherapy is a useful approach to improve long-term survival of patients with SCLC.
Conclusions
SCLC pathogenesis is a multi-step and multi-gene controlled process, involved in both genetic and epigenetic mechanisms. Aberrant alternation of TSGs silenced by promoter methylation is crucial in progression of SCLC, which has not been given well-deserved focus. Recently, more and more methylated TSGs have been identified in SCLC and thus provide a new insight into the mechanisms of initiation and development of SCLC. Methylated TSGs also provide new tumor makers, intervening approaches and prognostic factors for patients with SCLC. However, some problems remain to be solved. The key point is that CpG islands are located at various positions, such as exons and introns, not confined in promoter region of the TSGs. Although 79% of CpG islands are located in the promoter and the first exon, others are outside of these regions (19). Fukasawa M et al. (19) had reported most of detected TSGs were hyper-methylated in SCLC by microarray of cDNA library using CpG island clones. So data of methylation profile in promoter region is limited. The criteria of hyper-methylation is still unclear. Most of scholars define hyper-methylation of TSGs as follow: the ratio of normalized methylation intensity of cancer tissues to normalized methylation intensity of normal tissues is more than 3.0. The ratio less than 3.0 or less than background intensity represents un-methylation of TSGs. With the use of genome-wide epigenomic approaches (37) and scientific definition of hyper-methylation, the liable methylated TSGs might improve the early screen, treatment and prognosis of SCLC in future.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81172161).
Disclosure: The authors declare no conflict of interest.
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin 2008;58:71-96. [PubMed]
- Working Group Collaborated with IAP and IASLC. Tumours of the lung In: Travis WD, Brambilla E, Mueller-Hermelink HK, et al. eds. Tumours of the lung, pleura, thymus and heart. World Health Organization Classification of Tumours. Pathology & Genetics. Lyon: IARC Press, 2004:9-124.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [PubMed]
- Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res 2003;63:7089-93. [PubMed]
- Digel W, Lübbert M. DNA methylation disturbances as novel therapeutic target in lung cancer: preclinical and clinical results. Crit Rev Oncol Hematol 2005;55:1-11. [PubMed]
- Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science 2013;339:1567-70. [PubMed]
- Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell 1999;99:451-4. [PubMed]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33:245-54. [PubMed]
- Sameer AS, Abdullah S, Nissar S, et al. The blues of P(16)INK(4a): aberrant promoter methylation and association with colorectal cancer in the Kashmir valley. Mol Med Rep 2012;5:1053-7. [PubMed]
- Nyiraneza C, Sempoux C, Detry R, et al. Hypermethylation of the 5' CpG island of the p14ARF flanking exon 1β in human colorectal cancer displaying a restricted pattern of p53 overexpression concomitant with increased MDM2 expression. Clin Epigenetics 2012;4:9. [PubMed]
- Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225-9. [PubMed]
- Seike M, Gemma A, Hosoya Y, et al. Increase in the frequency of p16INK4 gene inactivation by hypermethylation in lung cancer during the process of metastasis and its relation to the status of p53. Clin Cancer Res 2000;6:4307-13. [PubMed]
- Sato M, Horio Y, Sekido Y, et al. The expression of DNA methyltransferases and methyl-CpG-binding proteins is not associated with the methylation status of p14(ARF), p16(INK4a) and RASSF1A in human lung cancer cell lines. Oncogene 2002;21:4822-9. [PubMed]
- Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta 2009;1796:114-28.
- Dammann R, Takahashi T, Pfeifer GP. The CpG island of the novel tumor suppressor gene RASSF1A is intensely methylated in primary small cell lung carcinomas. Oncogene 2001;20:3563-7. [PubMed]
- Pelosi G, Fumagalli C, Trubia M, et al. Dual role of RASSF1 as a tumor suppressor and an oncogene in neuroendocrine tumors of the lung. Anticancer Res 2010;30:4269-81. [PubMed]
- Grote HJ, Schmiemann V, Geddert H, et al. Methylation of RAS association domain family protein 1A as a biomarker of lung cancer. Cancer 2006;108:129-34. [PubMed]
- Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy 2006;2:74-9. [PubMed]
- Fukasawa M, Kimura M, Morita S, et al. Microarray analysis of promoter methylation in lung cancers. J Hum Genet 2006;51:368-74. [PubMed]
- Virmani A, Rathi A, Sugio K, et al. Aberrant methylation of TMS1 in small cell, non small cell lung cancer and breast cancer. Int J Cancer 2003;106:198-204. [PubMed]
- Fulda S, Küfer MU, Meyer E, et al. Sensitization for death receptor- or drug-induced apoptosis by re-expression of caspase-8 through demethylation or gene transfer. Oncogene 2001;20:5865-77. [PubMed]
- Pietanza MC, Kadota K, Huberman K, et al. Phase II trial of temozolomide in patients with relapsed sensitive or refractory small cell lung cancer, with assessment of methylguanine-DNA methyltransferase as a potential biomarker. Clin Cancer Res 2012;18:1138-45. [PubMed]
- Toyooka S, Toyooka KO, Maruyama R, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther 2001;1:61-7. [PubMed]
- Zöchbauer-Müller S, Minna JD, Gazdar AF. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist 2002;7:451-7. [PubMed]
- Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999;59:793-7. [PubMed]
- Zöchbauer-Müller S, Fong KM, Maitra A, et al. 5' CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res 2001;61:3581-5. [PubMed]
- Agathanggelou A, Dallol A, Zöchbauer-Müller S, et al. Epigenetic inactivation of the candidate 3p21.3 suppressor gene BLU in human cancers. Oncogene 2003;22:1580-8. [PubMed]
- Cohen AJ, Belinsky S, Franklin W, et al. Molecular and physiologic evidence for 5'CpG island methylation of the endothelin B receptor gene in lung cancer. Chest 2002;121:27S-28S. [PubMed]
- Guo M, Akiyama Y, House MG, et al. Hypermethylation of the GATA genes in lung cancer. Clin Cancer Res 2004;10:7917-24. [PubMed]
- Nishioka M, Kohno T, Tani M, et al. MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1, deleted, mutated, and methylated in human lung cancer. Proc Natl Acad Sci U S A 2002;99:12269-74. [PubMed]
- Kreisler A, Strissel PL, Strick R, et al. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 2010;29:5828-38. [PubMed]
- Oshita F, Sekiyama A, Suzuki R, et al. Detection of occult tumor cells in peripheral blood from patients with small cell lung cancer by promoter methylation and silencing of the retinoic acid receptor-beta. Oncol Rep 2003;10:105-8. [PubMed]
- Suzuki M, Shigematsu H, Shames DS, et al. Methylation and gene silencing of the Ras-related GTPase gene in lung and breast cancers. Ann Surg Oncol 2007;14:1397-404. [PubMed]
- Iwasaki Y, Sunaga N, Tomizawa Y, et al. Epigenetic inactivation of the thyroid hormone receptor beta1 gene at 3p24.2 in lung cancer. Ann Surg Oncol 2010;17:2222-8. [PubMed]
- Li JS, Ying JM, Wang XW, et al. Promoter methylation of tumor suppressor genes in esophageal squamous cell carcinoma. Chin J Cancer 2013;32:3-11. [PubMed]
- Zhang LX, Pan SY, Chen D, et al. Effect of adenomatous polyposis coli(APC) promoter methylation on gene transcription in lung cancer cell lines. Ai Zheng 2007;26:576-80. [PubMed]
- Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet 2008;24:231-7. [PubMed]


