The detectability of the pretreatment EGFR T790M mutations in lung adenocarcinoma using CAST-PCR and digital PCR
Introduction
Adenocarcinoma is the most common histological class of lung carcinoma, and its relative incidence is increasing (1). A great deal of progress has been made in targeted therapy for non-small cell lung cancer (NSCLC), largely as a result of the development of small-molecular inhibitors, such as epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) (2-4) and anaplastic lymphoma kinase (ALK) inhibitors (5). However, molecular targeted therapy is associated with some problems. A gatekeeper T790M mutation is known to cause resistance to EGFR-TKIs (6-10). This mutation is reported in 50–60% of cases with acquired resistance to EGFR-TKIs (7,9). The mutation detected rates are different from the mutation detection methods (11). T790M substitution has been shown to alter the proper binding of the drug to the ATP pocket of EGFR and/or return the affinity of the ATP to the drug to the wild-type level (8,9).
The mechanism through which patients develop acquired resistance remains to be elucidated. However, it has been reported that a small quantity of T790M mutation would be included in the pretreatment tumor of EGFR-TKIs (12-14). Watanabe et al. (12) reported that pretreatment EGFR T790M mutations were detected in NSCLC patients with EGFR activating mutations using a droplet digital PCR. However, they used genomic DNA (gDNA) extracted from formalin-fixed, paraffin-embedded (FFPE) samples. Thus, the poor preservation of the tumor tissues might have affected the reliability of their analysis.
In the present study, we analyzed the incidence and clinical significance of pretreatment T790M mutations in surgically resected lung adenocarcinoma tissues from tumors with EGFR-activating mutations using competitive allele-specific polymerase chain reaction (CAST-PCR) and a digital PCR. To increase the accuracy in the detection of T790M mutations, we used gDNA that had been extracted from frozen tumor specimens.
Methods
We studied 153 lung adenocarcinoma patients with EGFR-activating mutations who underwent surgery at Nagoya City University Hospital from 1997 to 2014. In all cases, EGFR-activating mutations were detected by the direct sequencing of EGFR (exon 18–21). In one case, we detected both L858R and T790M mutations in pretreated tissue specimens by direct sequencing (15). The characteristics of the 153 patients are shown in Table 1. Twenty-four of these patients were treated with EGFR-TKIs (either gefitinib or erlotinib). We used a CAST-PCR to examine the incidence of T790M mutations in all 153 patients. Twenty randomly selected samples were also subjected to a digital PCR. The present study was approved by the Institutional Review Board of Nagoya City University Hospital (No. 39), and written consent was obtained from all of the patients.
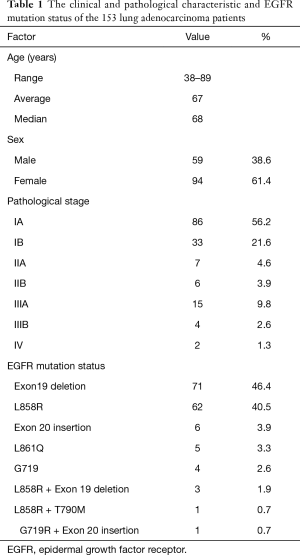
Full table
All of the tumor samples were immediately frozen and stored at −80 °C until use. Genomic DNA was extracted from tumor tissues using a Wizard SV Genomic DNA Purification System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The DNA concentration was determined using a NanoDrop spectrophotometer (NaoDrop Technologies, Ind. Rockland, DE, USA); the concentration was then adjusted to 20 ng/µL.
We used 4 µL of gDNA per sample for the PCRs to detect EGFR T790M mutation. The CAST-PCRs were run in a final volume of 20 µL in 96-well plates that included 10 µL of 2× TaqMan genotyping master mix (Life Technologies, Foster City, CA, USA), 2 µL of 10× Assay Mix, 5 µL of deionized water, and 4 µL of each sample. The CAST-PCRs were performed using a 7500 Fast Real-Time PCR System (Life Technologies), in accordance with the manufacturer’s protocol (15). The cycling conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 5 cycles at 92 °C for 15 sec and 58 °C for 1 min, 40 cycles at 92 °C for 15 sec and 60 °C for 1 min. The results of the mutation detection assays were analyzed using the Mutation DetectorTM Software version 2.0 (Life Technologies) and the %mutation value was determined. The %mutation value was calculated using the following formula:
%mutation = [1/2normalizedΔCt ÷ (1/2 normalizedΔCt + 1) ] ×100%
normalizedΔCt = [Ct (mutant allele assay) – Ct (wild type allele assay)] − CalibrationΔCtd
CalibrationΔCt = Ct (Mutant allele assay positive control) – Ct (Wild type allele)
We used 5 µL of gDNA per sample for the QuantStudioTM 3D Digital PCR assay. The digital PCR was run in a final volume of 15 µL, which included QuantStudioTM 3D Master Mix (Life Technologies) (7.5 µL), a 40× TaqMan Assay (EGFR_6240) 0.375 µL, nuclease-free water (2.125 µL), and 5 µL of each sample. The cycling conditions were as follows: initial denaturation at 96 °C for 10 min, followed by 39 cycles at 62 °C for 2 min, 98 °C for 30 sec and 60 °C for 2 min.
The overall survival (OS) and recurrence-free survival (RFS) of the 153 patients for whom a CAST-PCR was performed were assessed by the Kaplan-Meier method and differences were examined by the log-rank test. P values of <0.05 were considered to indicated statistical significance. All of the data were analyzed using the EZR software program (16).
Results
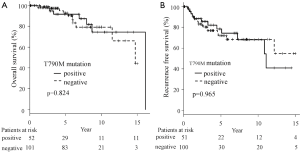
We used the CAST-PCR to detect T790M mutations in tissue specimens of 153 adenocarcinoma patients with EGFR-activating mutations. T790M mutations were detected in 45 (29.4%) of the 153 cases. The %mutation values of the 45 cases ranged from 0.13% to 2.65% (average 0.27%, median 0.20%). We analyzed the OS and RFS according to the T790M mutation status in the 153 cases (Figure 1A,B). T790M mutations did not have a significant effect on the OS or RFS. Furthermore, the prognosis of patients with or without T790M mutations did not differ to a statistically significant extent, irrespective of the status of the specimens before treatment. Although the results did not reach statistical significance, the patients without T790M mutations were likely to show a better prognosis than those with T790M mutations.
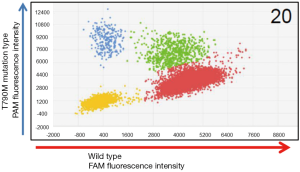
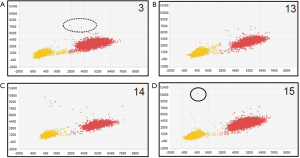
We also used a digital PCR assay to investigate the T790M mutation status of 20 of the 153 cases. The T790M mutation status was considered to be positive when at least one plot of T790M mutation-type high-intensity FAM fluorescence (blue plot) was present (Figures 2,3). T790M mutations that showed high-intensity FAM fluorescence and wild-type (green plot) were considered to be false positives—thus, the green plot was recognized as wild-type. Table 2 compares the results of the CAST-PCR and the digital PCR in the detection of EGFR T790M mutations. T790M mutations were detected in 8 out of the 20 (40%) cases in which mutations had been detected by the digital PCR. T790M mutations were detected in 15 of the 20 (75%) cases by the CAST-PCR.
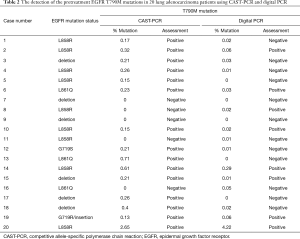
Full table
Discussion
Third-generation EGFR-TKIs that overcome EGFR-TKI-resistance in patients with T790M mutations have recently been developed. To prescribe these drugs to patients with acquired resistance, we need to perform re-biopsy to prove that a T790M mutation exists. This is associated with several problems. Re-biopsy is invasive, and, depending on the site of recurrence, it may be difficult to obtain a sufficient amount of tissue. The heterogeneity of the tumors can also affect the accuracy of the re-biopsy result. False-negative results therefore remain a possibility.
Thus, a method of detecting T790M mutations that are associated with resistance to treatment using either a small amount of tissue from the main tumor, or by analyzing tumor cells in blood, would be useful for identifying patients who are likely to acquire resistance to gefitinib or erlotinib. Third-generation EGFR-TKIs could be suitable for these patients.
The detection limit of direct sequencing is approximately 3%; thus, if the number of cells with T790M mutations is small, they would be undetectable by this technique. In our study, we used a CAST-PCR and a QuantStudioTM 3D Digital PCR to detect T790M mutations in such cases. Roma et al. (17) reported that it was possible to detect minimal EGFR mutations with high sensitivity and specificity using the CAST-PCR system. However, they concluded that the CAST-PCR was associated with a high false-positive rate in the detection of T790M mutations. On the other hand, Kinz et al. (18) reported that the QuantStudioTM 3D Digital PCR system was more sensitive than an allele-specific real-time quantitative polymerase chain reaction (RQ-PCR). Accurate quantitation revealed that the JAK2 V617F allele burden fell to 0.1%.
The use of the Cobas® EGFR mutation kit (Roche) with FFPE specimens is now acceptable for detecting EGFR mutations. We should investigate the appropriate samples and methods for determining the EGFR mutation status, which should be confirmed if EGFR inhibitors are to be properly administered. The identification of mutation-positive patients will allow patients who are more likely to benefit from molecular targeted drugs to be selected, while mutation-negative patients can avoid unnecessary side-effects associated with the use of molecular targeted drugs.
We should investigate methods for detecting small amounts of T790M mutations. We should also consider the conditions in which specimens are preserved. Based on this consideration, we used frozen tumor tissue specimens rather than FFPE tissue specimens. Our result concerning about T790M mutation detection rate is similar to some reports (19-21). On the other hands, some reports (22-25) are higher rate of T790M detection than our results. This discrepancy is considered to depend on the measurement method and the preservation condition of the specimens. When DNAs are damaged by the fixation of tumor tissues in formalin, artificial mutations, such as C-T or G-A transitions were detected in 90% of the samples. Thus, there were cases in which the detected T790M mutations were false-positives. In the present study, we detected T790M mutation in 29.4% of the pretreatment specimens with EGFR mutations (45 of 153 cases) using a CAST-PCR, and in 40% of pretreatment specimens with EGFR mutations (8 of 20cases) using a digital PCR. In contrast to our study, Watanabe et al. (12) and Iwama et al. (14) reported that they detected T790M mutations in 79.9% (298 of 373 cases) and 100% (25 of 25 cases) of pretreatment FFPE tissue specimens, respectively, using a digital PCR. Moreover, the incidence of pretreatment T790M mutations in the present study was lower in comparison to previous studies. The use of frozen, surgically-resected tissue specimens meant that our data was more precise. Thus, the false-positive rate would be expected to be lower in comparison to studies using FFPE tissue specimens. Moreover, our detection rate (for T790M mutations) was adjusted to the rate of acquired resistance (obtained from actual clinical data) (26,27). In interpreting the results of the reports, we should confirm the patients’ characteristics (including pathological stage), mutation detected method and condition of the tissues.
Previous studies (19-21) have indicated that the presence of the drug-resistance mutation T790M before initiation of treatment was associated with decreased PFS in patients with EGFR activating mutations who were receiving with gefitinib or erlotinib. In this study, almost patients were not received EGFR-TKIs. So, there is no difference about OS and RFS according to the T790M mutation status (Figure 1A,B).
It is necessary carefully consider patients with T790M mutations that are proven to confer EGFR-TKI resistance. The pretreatment tissue specimens obtained by re-biopsy had small T790M-resistant mutations, while some patients in whom the presence of T790M mutations was not detected in re-biopsy specimens, showed acquired resistance. We should establish new, reliable treatment strategies for patients with acquired resistant to EGFR-TKIs.
In the present study, we investigated the pretreatment EGFR T790M mutation status using frozen, surgically-resected tissue specimens from lung adenocarcinoma cases with EGFR-activating mutations using a digital PCR and a CAST-PCR. The mutation analysis showed that there was a discrepancy between their results. In order to clinically apply the T790M identification method using the pretreatment specimen for the patients with resistant to 1st or 2nd generation EGFR-TKI, it is necessary to establish an appropriate measurement method.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The present study was approved by the Institutional Review Board of Nagoya City University Hospital (No. 39), and written consent was obtained from all of the patients.
References
- Lewis DR, Check DP, Caporaso NE, et al. US Lung Cancer Trends by Histologic Type. Cancer 2014;120:2883-92. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Sasaki H, Shimizu S, Endo K, et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int J Cancer 2006;118:180-4. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786-792. [Crossref] [PubMed]
- Yano S, Yamada T, Takeuchi S, et al. Hepatocyte growth factor expression in EGFR mutant lung cancer with intrinsic and acquired resistance to tyrosine kinase inhibitors in a Japanese cohort. J Thorac Oncol 2011;6:2011-7. [Crossref] [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- Ohashi K, Maruvka YE, Michor F, et al. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol 2013;31:1070-80. [Crossref] [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [Crossref] [PubMed]
- Chen LY, Molina-Vila MA, Ruan SY, et al. Coexistence of EGFR T790M mutation and common activating mutations in pretreatment non-small cell lung cancer: A systematic review and meta-analysis. Lung Cancer 2016;94:46-53. [Crossref] [PubMed]
- Watanabe M, Kawaguchi T, Isa S, et al. Ultra-Sensitive Detection of the Pretreatment EGFR T790M Mutation in Non-small Cell Lung Cancer Patients with an EGFR-Activating Mutation Using Droplet Digital PCR. Clin Cancer Res 2015;21:3552-60. [Crossref] [PubMed]
- Isobe K, Hata Y, Tochigi N, et al. Usefulness of nanofluidic digital PCR arrays to quantify T790M mutation in EGFR-mutant lung adenocarcinoma. Cancer Genomics Proteomics 2015;12:31-7. [PubMed]
- Iwama E, Takayama K, Harada T, et al. Highly sensitive and quantitative evaluation of the EGFR T790M mutation by nanofluidic digital PCR. Oncotarget 2015;6:20466-73. [Crossref] [PubMed]
- Didelot A, Le Corre D, Luscan A, et al. Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR mutation detection in clinical formalin fixed paraffin embedded samples. Exp Mol Pathol 2012;92:275-80. [Crossref] [PubMed]
- Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical Statistics. Bone Marrow Transplant 2013;48:452-8. [Crossref] [PubMed]
- Roma C, Esposito C, Rachiglio AM, et al. Detection of EGFR mutations by TaqMan mutation detection assays powered by competitive allele-specific TaqMan PCR technology. Biomed Res Int 2013;13:385087.
- Kinz E, Leiherer A, Lang AH, et al. Accurate quantitation of JAK2 V617F allele burden by array-based digital PCR. Int J Lab Hematol 2015;37:217-24. [Crossref] [PubMed]
- Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol 2012;30:433-40. [Crossref] [PubMed]
- Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011;17:1160-8. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Williams C, Potén F, Moberg C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol 1999;155:1467-71. [Crossref] [PubMed]
- Quach N, Goodman MF, Shibata D. In vitro mutation artifacts after formalin fixation and error prone translesion synthesis during PCR. BMC Clin Pathol 2004;4:1. [Crossref] [PubMed]
- Ye X, Zhu ZZ, Zhong L, et al. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: truth or artifact? J Thorac Oncol 2013;8:1118-20. [Crossref] [PubMed]
- Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 2015;61:64-71. [Crossref] [PubMed]
- Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res 2011;17:1616-22. [Crossref] [PubMed]
- Rosell R, Dafini U, Felip E, et al. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicenter, single-arm, phase 2 trial. Lancet Respir Med 2017;5:435-44. [Crossref] [PubMed]


