Molecular biology of lung cancer
Introduction
The molecular basis of lung cancer is complex and heterogenous. Improvements in our understanding of molecular alterations at multiple levels (genetic, epigenetic, protein expression) and their functional significance have the potential to impact lung cancer diagnosis, prognostication and treatment. Lung cancers develop through a multistep process involving development of multiple genetic and epigenetic alterations, particularly activation of growth promoting pathways and inhibition of tumour suppressor pathways. Greater understanding of the multiple biochemical pathways involved in the molecular pathogenesis of lung cancer is crucial to the development of treatment strategies that can target molecular aberrations and their downstream activated pathways (1). Specific molecular alterations that drive tumour growth and provide targets for therapy have been best defined in adenocarcinomas (ADC) but there is increasing interest in the molecular landscape of squamous cell carcinoma (SCC) highlighting new potential therapeutic targets. In lung cancer as in other malignancies, tumourigenesis relates to activation of growth promoting proteins [e.g., v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), epidermal growth factor receptor (EGFR), BRAF, MEK-1, HER2, MET, ALK and rearranged during transfection (RET)] as well as inactivation of tumour suppressor genes [e.g., P53, phosphatase with tensin homology (PTEN), LKB-1] (1). Activation of growth promoting oncogenes can occur by gene amplification or other genetic alterations including point mutations and structural rearrangements leading to uncontrolled signalling through oncogenic pathways. “Oncogene addiction” results when cell survival depends on continued activation of the aberrant signalling (2,3) making them ideal candidates for targeted therapies. Oncogenic driver mutations have been identified in over 50% of lung ADC and are almost always exclusive of other driver mutations (4,5). Signalling pathways regulated by oncogenes and tumour suppressor genes are often interconnected with cross-talk between pathways involved in carcinogenesis. Added to the complexity is the occurrence of mutational evolution of tumours over time during the natural course of disease progression and in response to selection pressure exerted by therapy.
There is great genetic diversity in lung cancer and they harbour among the greatest numbers of genetic aberrations of all tumours (1). Understanding of the molecular biology of lung cancer has been revolutionised by next-generation sequencing technologies that provide a comprehensive means of identifying somatic alterations in entire cancer genomes or exomes. Lung cancers have highly complex genomes with a recent large-scale exome sequencing study of 31 non-small cell lung cancer (NSCLC) identifying 727 mutated genes not previously reported in the literature or in the COSMIC database (6). Genomic studies have confirmed previously well known alterations in lung cancer such as KRAS, EGFR and BRAF and have also identified low frequency but recurrent mutations that are novel in lung cancer (6-8) including potentially targetable alterations in JAK2, ERBB4 (8), RET (9-11), fibroblast growth factor receptor 1 (FGFR1) (12) and discoidin domain receptor 2 (DDR2) (13). While these studies provide a comprehensive portrait of genetic alterations in lung cancers, the challenge remains of identifying biologically relevant driver mutations from the vast majority of passenger mutations. The relative paucity of high frequency recurrent mutations highlights the heterogeneity and complexity of the molecular biology of lung cancer with common pathways affected by a range of different genetic alterations that poses a challenge for providing personalised medicine.
In this review, we discuss the most commonly altered and most clinically relevant oncogenes and tumour suppressor genes in lung cancer as improved understanding of the molecular pathology of lung cancer is crucial for advancements in treatment strategies.
KRAS
KRAS is part of the RAS family of proto-oncogenes (KRAS, NRAS and HRAS occurring in humans) and encodes a G-protein with a critical role in controlling signal transduction pathways which regulate cell proliferation, differentiation and survival (14). Ras proteins are guanosine diphosphate (GDP) bound and inactive in normal quiescent cells. There is a switch to the activated guanosine triphosphate (GTP) bound form following activation of upstream growth factor receptors. The activated Ras-GTP subsequently binds and activates a number of downstream pathways including mitogen-activated protein kinase (MAPK), RAS/RAF/MEK/MAPK pathway and the PI3-K [PI3K/AKT/mammalian target of rapamycin (mTOR)] pathways (15). KRAS plays a critical role in downstream signal transduction induced by a variety of growth factor receptors including EGFR and constitutive activation of the protein circumvents the need for growth factor mediated signalling. Activating mutations alter the GTPase activity of the protein hindering inactivation of the active RAS-GTP to GDP leading to increased signalling through multiple downstream growth promoting pathways (15). The RAS/RAF/MEK/MAPK signal transduction cascade plays a central role in many lung cancers with at least one mutations in the pathway identified in 132 of 188 tumours (7), of which the most common are mutations in KRAS.
Activating mutations in the KRAS oncogene are the commonest oncogenic alteration in lung ADC occurring in about 25-40% of cases (4,5,7,16-18) while HRAS and NRAS mutations are very rare (17). Differences in the prevalence of KRAS mutations in lung ADC most likely relate to different patient populations as KRAS mutations are more common in Western populations compared to Asian populations (19-22) and are more frequent in males and smokers (7,18,22). ADC in never smokers have been reported to harbour KRAS mutations in between 0-15% of cases (16,23). In addition, KRAS mutations are very rare or absent in SCCs and small cell cancer (17,24). Comprehensive genomic analysis of 188 SCCs identified only 1 KRAS mutation in codon 61 (12). KRAS mutations in lung adenocarcinoma consist of single amino acid substitutions in hotspots located mostly in codon 12 but also more rarely in codons 13 and 61 (14,17). The commonest mutations in KRAS are G to T transversions (~84%) in smokers while never smokers are more likely to harbour G to A transitions (16).
In keeping with the role of KRAS alterations as driver mutations, they do not occur in association with EGFR mutations (5,7,21,22), although rare exceptions do occur (18). A meta-analysis has shown KRAS mutant tumours are resistant to EGFR tyrosine kinase inhibitors (TKIs) (25), as KRAS mutations lead to constitutive activation of pathways downstream of EGFR. There is evidence that different KRAS mutant proteins have differing clinical significance. Interestingly, using data from the BATTLE trial (prospective phase II Biomarker-integrated Approaches of Targeted Therapy for Lung cancer Elimination), either G12C or G12V mutant KRAS predicted shorter progression free survival compared to other KRAS mutations or wild type KRAS (26). Furthermore, different amino acid substitutions were associated with activation of different pathways (PI3-K and MEK with Gly12Asp and Ral with Gly12Cys or mutant Gly12Val) resulting from divergent protein conformations from specific mutations leading to altered ability to associate with downstream protein mediators (26). This highlights that appropriate use of targeted therapies and clinical trial design needs to carefully evaluate the clinical and therapeutic significance of specific genetic alterations in lung cancer. The high frequency of KRAS mutations in lung cancer makes it an ideal therapeutic target but unfortunately clinical trials of targeted agents have generally been disappointing.
EGFR
Alterations of EGFR are involved in the pathogenesis of many tumours including NSCLC. EGFR encodes a transmembrane tyrosine kinase with an extracellular ligand-binding domain and an intracellular component including a tyrosine kinase domain (27). Binding of the ligand epidermal growth factor leads to receptor homo or heterodimerisation with other members of the EGFR family and activation of the tyrosine kinase domain (28,29). Signal transduction stimulated by EGFR occurs through the PI3K/AKT/mTOR, RAS/RAF//MAPK and JAK/STAT signalling pathways (28-30). EGFR is involved in regulation of numerous oncogenic functions such as cell proliferation, survival, differentiation, neovascularisation, invasion and metastasis (29,30). Activating mutations in EGFR lead to constitutive tyrosine kinase activation (30,31) and oncogenic transformation of lung epithelial cells in vitro (31). A transgenic mouse model with inducible expression of the commonest EGFR mutations showed development of multiple lung ADC that were sensitive to small molecule inhibition (32). Other mechanisms of increased EGFR signalling include increased protein expression or increased gene copy number (33,34).
Activating mutations of EGFR have been reported in 10-15% of unselected Western patients (5,21,35,36) and 30-40% of Asian populations (19,37,38). Differences in the reported prevalence rates of various mutations may in part relate to different patient populations but also depends on the sensitivity of mutation analysis techniques utilised in different studies. In NSCLC, EGFR mutations occur in the first four exons of the intracellular tyrosine kinase domain, most commonly exon 19 in frame deletions (~45%), of which there are over 20 variants, the commonest being delE746-A750. The next commonest EGFR mutations are missense mutations, particularly L858R, a single nucleotide point mutation in exon 21 leading to a single amino acid change from leucine to arginine at codon 858 (~40%). However, we found in an Australian population that exon 18 activating mutations constituted 14% of EGFR mutations in patients with early stage lung cancer and L858R mutations comprised only 29% of EGFR mutations present in this cohort (5). There are also a range of less common mutations including in frame duplications or insertions in exon 20 (~5-10%), of which there are many variants that are often associated with resistance to EGFR TKIs (22,39).
In lung cancer, almost all EGFR mutations occur in ADC (19,21,40,41) although they may also be seen in adenosquamous carcinomas. Mutations in EGFR are more commonly but not exclusively found in patients who are female, younger and with no history of smoking (7,19,21,22,37,40). EGFR mutations occur only very rarely, in histologically well sampled pure SCCs (24,42). However, comprehensive genomic analysis of 188 SCCs identified EGFR mutations in 2 cases, both with L861G mutations (12). While EGFR mutations are very rare in SCCs, variant-III mutations involving the extracellular domain of EGFR, copy-number gains and protein overexpression are more common in SCCs than in ADCs (43).
Secondary mutations in EGFR develop or are clonally selected in patients that develop resistance to EGFR TKIs, the commonest being the T790M activating point mutation in exon 20 which substitutes a “bulkier” methionine for threonine (44) that interferes with binding of reversible TKIs. T790M is found in about 50% of tumours from patients who develop acquired TKI resistance (41,44). Intriguingly, we observed that exon 20 mutations including T790M mutations associated with therapeutic resistance to EGFR TKI were seen in 29% of patients with EGFR mutations in a therapy naïve cohort (5). Activation of downstream pathways that bypass EGFR inhibition can also contribute to EGFR-TKI resistance including activation of PI3K pathway through amplification of MET (45).
BRAF
BRAF encodes a serine/threonine protein kinase that is the downstream effector protein of KRAS and activates the MAPK signal transduction pathway involved in regulation of cell proliferation and survival (46). Upon activation, BRAF phosphorylates downstream mediators MEK1 and MEK2 which subsequently activate ERK1 and ERK2, involved in regulation of growth regulating proteins such as c-JUN and ELK1 (14). Activating mutations in BRAF lead to increased kinase activity that exhibit transforming activity in vitro (46).
While activating BRAF mutations are common in melanoma (46), they occur in only about 3% of NSCLC (18,46-50). The mutations in NSCLC differ to those in melanoma and colorectal carcinoma with a lower proportion of V600E mutations that affect the kinase domain of the protein. In lung ADC, V600E mutations in exon 15 account for up to about 50% of BRAF mutations followed by G469A in exon 11 and D594G in exon 15 (48,50). Some of the BRAF mutations in NSCLC occur in the kinase domain (such as V600E, D594G and L596R) while others occur in the G-loop of the activation domain of the gene (such as G465V and G468A) (46). As BRAF and KRAS genes are part of the signalling pathway mediated by EGFR, it is not surprising that mutations in these genes are almost always mutually exclusive, in keeping with a common downstream pathway to transformation. BRAF mutations in lung cancer occur almost always in ADC (48). Non-V600E BRAF mutations have been associated with current or former smokers while V600E mutations appear to be more common in female never smokers (48,50). While uncommon, BRAF mutations represent an important therapeutic target due to the availability of targeted therapies already in clinical use for melanoma although there is only limited data about the clinical response to this approach in NSCLC (51).
MEK
MEK1 (also known as MAPK1) is a serine-threonine kinase that has an important function as a downstream target of RAS activation. MEK1 activates MAPK2 and MAPK3 downstream of BRAF (14). Rare cases of somatic mutations of MEK1 have been reported in NSCLC with 2 of 107 lung ADC found to have an activating mutation in exon 2 that did not involve the kinase domain (52). The mutations were exclusive of other driver mutations and were associated with gain of function in vitro (52).
MET
The proto-oncogene MET located on chromosome 7q21-q31 encodes a membrane tyrosine kinase receptor that is also known as hepatocyte growth factor receptor (53). Upon binding of its ligand hepatocyte growth factor, there is receptor homodimerisation, kinase activation and signalling through downstream pathways including RAS/RAF/MEK/MAPK, PI3K/AKT and c-SRC kinase pathways (53). In NSCLC, MET is altered by gene amplification in about 1-7% of treatment naive patients (54-57) but in one study amplification was found in 21% of patients (58). Increased MET copy number may be more common in SCC than ADC (57) and is mutually exclusive with KRAS mutations (56,58). MET amplification results in overexpression of MET protein and activation of downstream signal transduction pathways. The oncogenic activity of MET has been demonstrated in vitro with evidence of gene amplification associated with constitutive receptor phosphorylation, activation of the PI3K/AKT pathway and sensitivity to MET inhibition (45,59). Amplification of MET is a known mechanism of secondary EGFR-TKI resistance with this kinase switch occurring in approximately 20% of patients with acquired resistance (45,54,55). In this scenario, MET amplification drives and maintains the PI3K/AKT pathway bypassing EGFR blockade by TKIs (45), suggesting concomitant MET inhibition may be a means of overcoming TKI resistance. Mutations of MET also occur uncommonly in about 3-5% of ADC (7,56).
HER2
The human epidermal growth factor receptor 2 (HER2/ERBB2) gene encodes a membrane bound receptor tyrosine kinase that is a member of the ERBB family of receptors, along with EGFR. Unlike other ERBB receptors, it does not bind ligand directly but can form heterodimers with other ligand-bound members of the receptor family (60). Activation leads to signalling through a variety of signal transduction pathways including PI3K, MAPK and JAK/STAT pathways (61). Activation of HER2 occurs in a small proportion of lung cancers with overexpression in approximately 20% of cases, gene amplification in 2% (62) and activating mutations in 1.6-4% of NSCLC (63-65). Activating mutations of HER2 are exon 20 in frame insertions of 3 to 12 base pairs in length (63). There is in vivo evidence of the oncogenic activity of HER2 with multiple adenosquamous carcinomas developing in a transgenic mouse model expressing mutant HER2 and exhibiting susceptibility to small molecule inhibition (66). Alterations of HER2 occur mostly in ADC (63-65) and mutations occur in tumours that are wild-type for EGFR and KRAS (63,64) and in some studies, are associated with female gender, Asian ethnicity and non-smoking status (63,65), similar to the clinical profile of EGFR mutant tumours.
PI3K/AKT/mTOR
The PI3K/AKT/mTOR pathway is an important signal transduction pathway involved in regulation of cell proliferation, survival, differentiation adhesion and motility (67,68). Alterations of this pathway have been implicated in both NSCLC and small cell carcinoma (69,70). The pathway is activated through activation of a variety of membrane tyrosine kinase receptors including EGFR, HER2, insulin-like growth factor receptor, vascular endothelial growth factor receptor and platelet derived growth factor receptor (71,72). Activated receptor tyrosine kinases recruit PI3K to the cell membrane where it phosphorylates PIP2 to PIP3 [phosphatidylinositol 4,5-bisphosphate (PIP2) to phosphatidylinositol 3,4,5-triphosphate]. PIP3 in turn recruits the serine threonine kinase AKT to the membrane where it is phosphorylated by 3-phosphoinositide-dependent kinase 1 (PI3 kinase) and mTOR. mTOR is a serine/threonine kinase that is a downstream target of AKT (72). Activated AKT in turn activates multiple targets including tuberous sclerosis 2 and Bcl-2 associated death promotor leasing to cell proliferation and survival [reviewed in (71)]. There is also interaction with other pathways including RAS/RAF/MEK (Rat sarcoma/rapidly accelerated fibrosarcoma/MAPK or Erk kinase) with RAS having the capacity to directly activate PI3K (72).
The PI3K/AKT/mTOR pathway is frequently deregulated in many tumours including 50-70% of NSCLC (7,71). Significant alterations involving the PI3K pathway were identified in 47% of SCCs in the Cancer Genome Atlas project (12). Pathway activation in lung carcinogenesis occurs through a variety of mechanisms including activating mutations in EGFR, KRAS, PI3K or AKT (68,71) as well as PIK3CA amplification, or loss of negative regulation by the tumour suppressor gene PTEN (72).
The PI3K protein family (phosphatidylinositol 3-kinases) are intracellular lipid kinases and the main catalytic subunit, the p110alpha isoform, is encoded by the PIK3CA gene (71). Activating mutations and amplification of PIK3CA cause constitutive ligand-independent pathway activation (73,74). PIK3CA mutations mostly involve the catalytic domain and have been identified in approximately 1-3% of NSCLCs (7,73,75) and are more common in SCC than ADCs (4,75). Unlike most oncogenic driver mutations, PIK3CA mutations may occur in association with EGFR or KRAS mutations (5,73,75) suggesting they may not represent true driver mutations. However, in vitro studies of lung cancer cell lines with PIK3CA mutations or copy number gains show increased PI3 kinase activity sensitive to small molecule inhibition (73) and in vivo mouse models with PIK3CA mutation expression develop numerous ADC, suggesting oncogenic activity (74). PIK3CA may also be amplified in NSCLC, especially in SCCs (73,76) and increased copy number of PIK3CA has been reported in ~5% of small cell carcinoma cell lines (73). Although rare, PI3K/AKT/mTOR pathway activation can also occur through AKT mutations which have been reported in 0.5-2% of NSCLC (5,7,77), particularly SCCs (77).
ALK
Rearrangements of the receptor tyrosine kinase ALK resulting most commonly in fusions of the intracellular kinase domain with the amino terminal end of echinoderm microtubule associated protein-like 4 (EML4) occur in a subset of lung cancers (78-80). The rearrangement results from a short inversion in chromosome 2p, whereby in the commonest variant, intron 13 of EML4 is fused to intron 19 of ALK {ALK [inv (2) (p21; p23)]} (79). Numerous variants of EML4-ALK fusions have been identified due to differing lengths of EML4, the commonest being exons 1-13 of EML4 joining to exons 20-29 of ALK (78,81,82). More recently, different partner genes have been identified in a small subset of ALK rearrangements (<1% of cases) including KIF5B (kinesin family member 5b), TFG (TRK -fused gene) and KLC-1 (kinesin light chain1) (83,84). The oncogenic EML4-ALK fusion protein has a constitutively activated kinase and has gain of function activity in vitro (80) and in vivo mouse models expressing EML4-ALK develop multiple lung ADC that are susceptible to pharmacologic ALK inhibition (85). Activation of ALK is linked to cell proliferation and inhibition of apoptosis mediated through the RAS/RAF/MAPK1, PI3K/AKT and JAK3-STAT3 signalling pathways (82).
ALK rearrangements have been identified in approximately 4% of unselected NSCLC (86) although some studies have found a slightly lower prevalence (5,87). They are more commonly found in ADC from younger patients who are never smokers or light smokers (78,87-91) and almost always occur in ADCs (90). While ALK rearrangements are usually mutually exclusive with EGFR and KRAS mutations (5,87,91,92) cases of coexistent EGFR mutations have been reported and provide a mechanism for TKI resistance (78,93-95). While ALK inhibition with the tyrosine kinase inhibitor crizotinib produces profound responses, drug resistance develops with evidence of secondary ALK point mutations and activation of EGFR signalling implicated in some cases (81,93).
ROS1
ROS1 is a proto-oncogene located on chromosome 6q22 which encodes a transmembrane tyrosine kinase receptor that has high homology with ALK in its protein kinase domain (96). ROS1 activation leads to signalling through the PI3K/AKT/mTOR, STAT3 and RAS/MAPK/ERK pathways (96). In 2007, a large scale phosphoproteomic screen for tyrosine kinase activity in lung cancer identified ROS1 fusion in a NSCLC cell line (1 of 41) and a patient sample (1 of 150) (SLC34A2-ROS1 and CD74-ROS1 respectively) (83). Subsequently, a novel KDELR2-ROS1 in-frame fusion was identified in an adenocarcinoma from a non-smoker using whole genome and transcriptome sequencing (8). In 2 large studies using FISH, ROS1 rearrangements were found in 18 of 694 ADCs (2.6%) (97) and 13 of 1,116 ADCs (1.2%) (98). A variety of 5' fusion partners have been identified in ROS1 gene rearrangements (including FIG, KDELR2, TPM3, SDC4, LRIG3, EZR, SLC34A2 and CD74) and it is uncertain what role, if any, the partner plays in the oncogenic function of the fusion kinase (8,83,98). Interestingly, ROS1 rearrangements appear to be more common in patients who are younger, never smokers or of Asian ethnicity (97) similar to ALK rearrangements (90). Furthermore, there is in vitro and early clinical evidence that lung cancers with ROS1 rearrangements are sensitive to kinase inhibitors including the ALK/MET inhibitor crizotinib (97).
RET
RET is located on chromosome 10q11.2 and encodes a receptor tyrosine kinase involved in neural crest development. Alterations of RET have long been known to play a role in papillary and medullary thyroid carcinoma (99) but it was not until recently that activation of RET through chromosomal rearrangement has been identified in a small proportion of lung cancers (9-11). The translocation fuses the functional RET kinase domain from exons 12-20 to KIF5B (kinesin family 5B gene), that is 10Mb from RET on chromosome 10 and encodes a coiled coil domain involved in organelle trafficking (9,10). KIF5B-RET fusions have been identified in 1-2% of lung ADC using massively parallel sequencing technologies (10,11) and to date have been found to be mutually exclusive of other driver mutations involving EGFR, KRAS or ALK. In a highly selected cohort of lung ADC from never smokers or light smokers known to be wild type for other driver mutations (EGFR, KRAS, ALK, HER2, BRAF and ROS1), 10 of 159 (6.3%) harboured RET rearrangements (11). Similar to ALK and ROS1, rearrangements of RET also appear to be associated with ADC from never smokers (9-11). Importantly, there are several multi-kinase inhibitors that are effective against RET and there is in vitro evidence that cell lines expressing KIF5B-RET fusions are sensitive to RET inhibition (10,11).
FGFR1
Somatic gene amplifications have been found in SCCs in a number of genes including SOX, PDGFRA (12) and FGFR1 (12,100). FGFR1 is a membrane receptor tyrosine kinase that regulates cell proliferation through activation of the MAPK and PI3K pathways (101). Amplification of FGFR1 has an oncogenic effect on NSCLC cell lines in vitro that is sensitive to small molecule inhibition (102). About 20% of SCCs have been shown to harbour FGFR1 amplifications but the abnormality is uncommon in ADCs (100,102).
DDR2
Recently, a sequencing screen including the entire tyrosine kinome was undertaken in SCCs and mutations were identified in DDR2 in 3.8% of cases (13). DDR2 encodes a membrane-bound receptor tyrosine kinase that binds collagen and is involved in regulation of cell proliferation and survival (103). Mutations of DDR2 are associated with oncogenic activity in vitro that is sensitive to inhibition with dasatinib (13).
Tumour suppressor genes
Tumour suppressor genes are crucial negative regulators of normal cell growth. Loss of tumour suppressor gene (TSG) function is an important mechanism of carcinogenesis and requires inactivation of both gene alleles, as outlined in Knudson’s two hit hypothesis (104). In one allele, the individual gene is often inactivated by mutation, epigenetic silencing or other aberrations, while the second allele is often inactivated through loss of heterozygosity (LOH) whereby a region of the chromosome is lost by deletion, nonreciprocal translocation or mitotic recombination. In lung cancer, TSGs that are frequently inactivated include TP53, retinoblastoma 1 (RB1), serine-threonine kinase 11 (STK11), CDKN2A, FHIT, RASSF1A and PTEN (1,7,105) and these genes map to chromosomal regions commonly identified in LOH studies. For example, regions frequently exhibiting allelic loss in lung cancer involve known TSGs such as TP53 (17p13), RB (13q12), p16 (9p21), and PTEN (10q22) (105). In a study by Ding et al. (7), mutations were identified in several TSGs not previously known to play a significant role in lung adenocarcinoma including the TSG NF1 (involved in neurofibromatosis type 1), that was mutated in 13 tumours and the TP53 regulator ATM in 13 patients.
TP53
TP53 located on chromosome 17p13 encodes a nuclear phosphoprotein of 53 kDa that identifies and binds to regions of damaged DNA (106) and acts as a transcription factor controlling the expression of a multitude of different genes. Damaged DNA or carcinogenic stress induces TP53 leading to cell cycle arrest by inducing expression of cyclin dependent kinase inhibitors to enable DNA repair or apoptosis. TP53 inactivation is one of the most significant genetic abnormalities in lung cancer with hemizygous deletion of 17p13, containing the locus of TP53, occurring in 90% of small cell carcinomas and about 65% of NSCLC (107). Inactivating mutations in TP53 (mostly missense mutations within the DNA-binding domain) have been reported in 80-100% of small cell lung carcinomas (108). By contrast, a meta-analysis of TP53 in over 4,000 NSCLC found alterations by mutation or protein accumulation in only 46.8% of cases (109), more commonly in SCC than ADC and associated with higher tumour stage, grade and male gender. Mutations of TP53 were found in at least 81% of SCCs that underwent comprehensive genomic analysis as part of The Cancer Genome Atlas (TCGA) project (12). Ding et al. (7) found TP53 mutations in 85 of 188 ADC (45%). In NSCLC, TP53 mutations are associated with a positive smoking history or exposure to environmental tobacco smoke (19,110). The mutational spectrum of different types of TP53 mutations also differs between smokers and non-smokers with smoking related cancers having a significantly higher frequency of G to T transversions compared to G to C transversions (thought to be induced by polycyclin aromatic hydrocarbons in tobacco smoke) and G to A transitions at CpG dinucleotides more commonly seen in never smokers (110,111). A meta-analysis of 74 studies showed that aberrant p53 detected by protein expression or mutational analysis is an unfavourable prognostic factor in lung NSCLC (112). Genetic alterations of TP53 have also been associated with treatment resistance (106). TP53 gene mutations can occur in association with EGFR and KRAS mutations (19).
PTEN
PTEN encodes a lipid and protein phosphatase on chromosome 10 that inhibits the PI3K/AKT/mTOR signalling pathway by dephosphorylating PI-(3,4,5)-triphosphate (68). Inactivation of the TSG function of PTEN leads to unrestricted activation of AKT/protein kinase B independent of ligand binding (68). Mutations of PTEN occur only rarely in about 5% of NSCLC (113) being more common in SCC than ADC (10.2% vs. 1.7%) and associated with a history of smoking. By contrast, reduced protein expression has been reported in about 75% of NSCLC (114).
LKB1 (STK11)
LKB1 (also known as STK11) is a TSG located on chromosome 19p13 that encodes a serine-threonine kinase that inhibits mTOR and has been implicated in a range of biological processes including regulation of the cell cycle, chromatin remodelling, cell polarity, and energy metabolism (115,116). Deregulation of mTOR pathway components (not including KRAS mutations) has been reported in 30% of ADCs (7). Germline mutations of LKB1/STK11 occur in patients with Peutz-Jeghers syndrome (115). In lung cancer, LKB1 may be inhibited by a variety of somatic mutations or deletions that produce truncated proteins with inactivation of LKB1 occurring in about 11-30% of lung ADC (7,117-119), making it the third commonest genetic aberration in lung ADC after TP53 and KRAS. LKB1 inactivation is more common in lung ADC compared to SCCs (117,119). There is some evidence of an association between LKB1 mutations and a history of smoking (117) in men (118,120) and a correlation with KRAS mutations has also been reported (117,118).
The p16INK4a-cyclin D1-CDK4-RB pathway
The p16INK4A/RB pathway regulates cell cycle progression from G1 to S phase. RB1 is a tumour suppressor gene that encodes RB protein which regulates cell cycle G1/S transition by binding the transcription factor E2F1. RB1 was the first TSG described in lung cancer (121) and is inactivated in about 90% of small cell lung carcinomas but only about 10-15% of NSCLC (1). In NSCLC, the pathway is mostly switched off through alterations of cyclin D1, CDK4 and the cyclin dependent kinase inhibitor p16 (CDKN2A) (105). p16INK4A inhibits cyclin D1 dependent phosphorylation of RB protein, thereby preventing cell cycle transition through the G1/S checkpoint (122). p16INK4A is inactivated in about 80% of NSCLC (123,124) and was altered in 72% of lung SCCs examined by TCGA, mostly through homozygous deletion, methylation or inactivating mutations (12). In addition, there is overexpression of cyclin D1 through gene amplification or other mechanisms in about 40% of NSCLC (123).
Molecular targeting in NSCLC
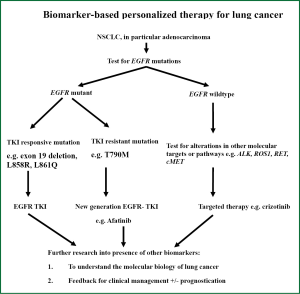
The presence of these molecular targets as described above now defines the characteristics of NSCLC, with EGFR mutation and ALK rearrangements being the most clinically relevant at present (125). The prevalence of these mutations varies in lung cancer arising from patient in different regions (126). Activating EGFR mutations were found in up to 20% of Caucasians while in the Asian populations these EGFR mutations can be present in up to 40% of patients with NSCLC (127). These ethnic difference in NSCLC properties appears to be not limited to the presence of activating EGFR mutations but is also evident in other driver oncogenic mutation profiles (including ALK, KRAS, MET etc.), histology and hence tumour response to targeted therapy treatment (63,126,128). The presence of these driver mutations is generally found to be mutually exclusive to others in the same tumour (126). In lung ADC among Asians, ALK rearrangement is seen in up to 7% of patients with lung ADC (79). Lung tumours bearing EML4-ALK rearrangement are non-responsive to conventional chemotherapy or EGFR-tyrosine kinase inhibitors but are sensitive to a specific tyrosine kinase inhibitor named crizotinib (129). Based on our current understanding of therapeutic molecular targets of EGFR mutation and ALK gene rearrangement in NSCLC and the availability of corresponding targeted agents, an algorithm of testing for molecular targets in NSCLC is proposed as in Figure 1, which represents a stepwise approach to testing for individual targets, beginning with EGFR then, if negative, ALK fusion gene or other potential targets if appropriate.
Among NSCLC, adenocarcinoma accounts for up to 80% of histological subtypes (130). There are previous reports of correlations between histological subtypes of ADC demonstrating micropapillary features with presence of activating EGFR mutations, leading to the suggestions that the presence of specific mutations in NSCLC actually represent heterogeneity in cancer biology and also response to therapy (131). Given the heterogeneity of lung cancer histology, however, histological subtypes are difficult to be used as the sole reliable marker for guidance to molecular phenotyping and selection of targeted therapy (132,133).
Targeting therapeutic oncogenic mutations like EGFR and ALK can give dramatic initial treatment response or at least an initial stable clinical disease. The response rate is up to 70% in lung ADC bearing favourable activating EGFR mutations (134). The median progression free survival is usually quoted as 9-11 months with different tyrosine kinase inhibitors (135,136), after which most patients with EGFR mutations will experience disease progression and drug resistance. A proportion of such drug resistance is attributed to the development of a second mutation, usually T790M at exon 20 (137). It is hard to explain the eventual loss of drug sensitivity in tumours bearing those favourable EGFR mutations (exon 19 deletions and L858R) even without the acquisition of secondary mutations like T790M or the presence of other uncommon or less favourable EGFR mutations. This could reflect suboptimal therapeutic targeting and better understanding on the biology of EGFR-related tumour signalling and other oncogenic mutations will improve drug targeting and give patients better prediction of therapeutic response and prognostication.
Conclusions
The identification of driver mutations in EGFR and ALK heralded a new era of targeted therapy in lung adenocarcinoma and advanced sequencing technologies are providing even more sophisticated insights into the molecular aberrations in oncogenes and tumour suppressor genes underlying lung cancer (12,138-142). These studies have identified a range of potentially targetable genetic aberrations in lung cancer but have also highlighted a troubling complexity and heterogeneity which poses significant challenges for molecular diagnosis and targeted treatment. Greater knowledge of the molecular biology and genomic landscape of lung cancer offers promise for the future. Improvements in outcome from lung cancer will almost certainly require the identification of increasing numbers of ever rarer driver mutations, and diagnostic approaches that can identify multiple therapeutic targets offer significant advantages. However, the identification of driver genomic aberrations also requires the parallel development of effective targeted therapies and for many of these changes (such as KRAS) such therapies are not yet available. Resistance to targeted therapeutics is an increasingly recognised issue into which genomic analyses may provide important mechanistic insights underlying future rational therapeutic approaches.
Acknowledgements
Funding sources: Sydney Foundation for Medical Research; Hong Kong SK Yee Medical Foundation; Cancer Institute NSW Clinical Research Fellowship 10/1/07; Sydney Breast Cancer Foundation; Lifehouse at Royal Prince Alfred Hospital Grant; Lung Cancer SPORE P50CA70907.
Disclosure: WC has received honoraria from Pfizer Oncology. SOT is a member of the Roche Australia Molecular Pathology Advisory Board. The authors declare no other conflicts of interest
References
- Larsen JE, Minna JD. Molecular biology of lung cancer: clinical implications. Clin Chest Med 2011;32:703-40. [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [PubMed]
- Weinstein IB. Cancer. Addiction to oncogenes-the Achilles heal of cancer. Science 2002;297:63-4. [PubMed]
- Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24. [PubMed]
- Yip PY, Yu B, Cooper WA, et al. Patterns of DNA mutations and ALK rearrangement in resected node negative lung adenocarcinoma. J Thorac Oncol 2013;8:408-14. [PubMed]
- Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole exome sequencing. Carcinogenesis 2012;33:1270-6. [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [PubMed]
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [PubMed]
- Ju YS, Lee WC, Shin JY, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 2012;22:436-45. [PubMed]
- Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. [PubMed]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [PubMed]
- Hammerman PS, Sos ML, Ramos AH, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov 2011;1:78-89. [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [PubMed]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol 2008;9:517-31. [PubMed]
- Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731-4. [PubMed]
- Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res 1992;52:2665s-9s. [PubMed]
- Schmid K, Oehl N, Wrba F, et al. EGFR/KRAS/BRAF mutations in primary lung ADC and corresponding locoregional lymph node metastases. Clin Cancer Res 2009;15:4554-60. [PubMed]
- Kosaka T, Yatabe Y, Endoh H, et al. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004;64:8919-23. [PubMed]
- Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung cancer 2010;69:272-8. [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [PubMed]
- Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non–small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res 2006;12:1647-53. [PubMed]
- Subramanian J, Govindan R. Molecular genetics of lung cancer in people who have never smoked. Lancet Oncol 2008;9:676-82. [PubMed]
- Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res 2012;18:1167-76. [PubMed]
- Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer Lancet Oncol 2008;9:962-72. [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS Oncogene Substitutions on Protein Behavior: Implications for Signaling and Clinical Outcome. J Natl Cancer Inst 2012;104:228-39. [PubMed]
- Prenzel N, Fischer OM, Streit S, et al. The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr Relat Cancer 2001;8:11-31. [PubMed]
- Scagliotti GV, Selvaggi G, Novello S, et al. The Biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res 2004;10:4227s-32s. [PubMed]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127-37. [PubMed]
- Sordella R, Bell DW, Haber DA, et al. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004;305:1163-7. [PubMed]
- Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med 2005;2:e313. [PubMed]
- Ji H, Li D, Chen L, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell 2006;9:485-95. [PubMed]
- Dahabreh IJ, Linardou H, Siannis F, et al. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non–small cell lung cancer. Clin Cancer Res 2010;16:291-303. [PubMed]
- Okabe T, Okamoto I, Tamura K, et al. Differential constitutive activation of the epidermal growth factor receptor in non–small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res 2007;67:2046-53. [PubMed]
- Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in non–small-cell lung cancer working group: standardization for use in the clinical trial setting. J Clin Oncol 2008;26:983-94. [PubMed]
- Russell PA, Barnett SA, Walkiewicz M, et al. Correlation of mutation status and survival with predominant histologic subtype according to the new IASLC/ATS/ERS lung adenocarcinoma classification in stage III (N2) patients. J Thorac Oncol 2013;8:461-8. [PubMed]
- Tokumo M, Toyooka S, Kiura K, et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non–small cell lung cancers. Clin Cancer Res 2005;11:1167-73. [PubMed]
- Yoshida K, Yatabe Y, Park JY, et al. Prospective validation for prediction of gefitinib sensitivity by epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer. J Thorac Oncol 2007;2:22-8. [PubMed]
- Yamamoto H, Toyooka S, Mitsudomi T. Impact of EGFR mutation analysis in non-small cell lung cancer. Lung cancer 2009;63:315-21. [PubMed]
- Marchetti A, Ardizzoni A, Papotti M, et al. Recommendations for the analysis of ALK gene rearrangements in non–small-cell lung cancer: a consensus of the Italian Association of Medical Oncology and the Italian Society of Pathology and Cytopathology. J Thorac Oncol 2013;8:352-8. [PubMed]
- Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res 2008;14:4877-82. [PubMed]
- Ohtsuka K, Ohnishi H, Fujiwara M, et al. Abnormalities of epidermal growth factor receptor in lung squamous-cell carcinomas, adenosquamous carcinomas, and large-cell carcinomas: tyrosine kinase domain mutations are not rare in tumors with an adenocarcinoma component. Cancer 2007;109:741-50. [PubMed]
- Heist RS, Sequist LV, Engelman JA. Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol 2012;7:924-33. [PubMed]
- Balak MN, Gong Y, Riely GJ, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor–mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res 2006;12:6494-501. [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [PubMed]
- Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 2002;62:6997-7000. [PubMed]
- Marchetti A, Chen TH, Richards WG, et al. Clinical features and outcome of patients with non–small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [PubMed]
- Naoki K, Chen T, Richards W, et al. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res 2002;62:7001-3. [PubMed]
- Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol 2011;29:2046-51. [PubMed]
- Falchook GS, Long GV, Kurzrock R, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet 2012;379:1893-901. [PubMed]
- Marks JL, Gong Y, Chitale D, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signalling pathwar genes in lung adenocarcinoma. Cancer Res 2008;68:5524-8. [PubMed]
- Sadiq AA, Salgia R. MET as a possible target for non–small-cell lung cancer. J Clin Oncol 2013;31:1089-96. [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA 2007;104:20932-7. [PubMed]
- Cappuzzo F, Marchetti A, Skokan M, et al. MET increased gene copy number negatively affects survival of surgically resected non–small-cell lung cancer patients. J Clin Oncol 2009;27:1667-74. [PubMed]
- Onozato R, Kosaka T, Kuwano H, et al. Activtion of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009;4:5-11. [PubMed]
- Go H, Jeon YK, Park HJ, et al. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol 2010;5:305-13. [PubMed]
- Beau-Faller M, Ruppert AM, Voegeli AC, et al. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J Thorac Oncol 2008;3:331-9. [PubMed]
- Lutterbach B, Zeng Q, Davis LJ, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res 2007;67:2081-8. [PubMed]
- Tzahar E, Waterman H, Chen X, et al. A hierarchical network fo interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol 1996;16:5276-87. [PubMed]
- Graus-Porta D, Beerli RR, Daly JM, et al. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J 1997;16:1647-55. [PubMed]
- Heinmöller P, Gross C, Beyser K, et al. HER2 status in non-small cell lung cancer: results from patient screening for enrollment to a phase II study of herceptin. Clin Cancer Res 2003;9:5238-43. [PubMed]
- Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005;65:1642-6. [PubMed]
- Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature 2004;431:525-6. [PubMed]
- Tomizawa K, Suda K, Onozato R, et al. Prognostic and predictive implications of HER2/ERBB2/neu gene mutations in lung cancers. Lung Cancer 2011;74:139-44. [PubMed]
- Perera SA, Li D, Shimamura T, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci 2009;106:474-9. [PubMed]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006;7:606-19. [PubMed]
- Cully M, You H, Levine AJ, et al. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer 2006;6:184-92. [PubMed]
- Brognard J, Clark AS, Ni Y, et al. Akt/Protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res 2001;61:3986-97. [PubMed]
- Tsurutani J, West KA, Sayyah J, et al. Inhibition of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathway but not the MEK/ERK pathway attenuates laminin-mediated small cell lung cancer cellular survival and resistance to imatinib mesylate or chemotherapy. Cancer Res 2005;65:8423-32. [PubMed]
- Papadimitrakopoulou V. Development of PI3K/AKT/MTOR pathway inhibitors and their application in personalized therapy for non-small-cell lung cancer. J Thorac Oncol 2012;7:1315-26. [PubMed]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase-AKT pathway in human cancer. Nat Rev Cancer 2002;2:489-501. [PubMed]
- Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res 2008;68:6913-21. [PubMed]
- Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant K-ras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351-6. [PubMed]
- Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung cancer 2006;54:209-15. [PubMed]
- Ji M, Guan H, Gao C, et al. Highly frequent promoter methylation and PIK3CA amplification in non-small cell lung cancer (NSCLC). BMC Cancer 2011;11:147. [PubMed]
- Malanga D, Scrima M, De Marco C, et al. Activating E17K mutation in the gene encoding the protein kinase AKT1 in a subset of squamous cell carcinoma of the lung. Cell Cycle 2008;7:665-9. [PubMed]
- Koivunen JP, Mermel C, Zejnullahu K, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008;14:4275-83. [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [PubMed]
- Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoform of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res 2008;68:4971-6. [PubMed]
- Choi YL, Soda M, Yamashati Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [PubMed]
- Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res 2011;17:2081-6. [PubMed]
- Rikova K, Guo A, Zeng Q. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [PubMed]
- Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res 2009;15:3143-9. [PubMed]
- Soda M, Takada S, Takeuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci USA 2008;105:19893-7. [PubMed]
- Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements. A new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol 2009;4:1450-4. [PubMed]
- Selinger CI, Rogers TM, Russell PA, et al. Testing for ALK rearrangement in lung adenocarcinoma-a multicenter comparison of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol 2013. [Epub ahead of print]. [PubMed]
- Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the Western population. Clin Cancer Res 2009;15:5216-23. [PubMed]
- Sakairi Y, Nakajima T, Yasufuku K, et al. EML4-ALK fusion gene assessment using metastatic lymph node samples obtained by endobronchial ultrasound-guided transbronchial needle aspiration. Clin Cancer Res 2010;16:4938-45. [PubMed]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247-53. [PubMed]
- Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 2009;115:1723-33. [PubMed]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009;22:508-15. [PubMed]
- Sasaki T, Koivunen J, Ogino A, et al. A novel ALK secondary mutation and EGFR signalling cause resistance to ALK kinase inhibitors. Cancer Res 2011;71:6051-60. [PubMed]
- Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal etection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol 2013;8:322-8. [PubMed]
- Tiseo M, Gelsomino F, Boggiani D, et al. EGFR and EML4-ALK gene mutations in NSCLC: A case report of erlotinib-resistant patient with both concomitant mutations. Lung cancer 2011;71:241-3. [PubMed]
- Chin LP, Soo RA, Soong R, et al. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non–small-cell lung cancer. J Thorac Oncol 2012;7:1625-30. [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nature Med 2012;18:378-81. [PubMed]
- Wells SA Jr, Santoro M. Targeting the RET pathway in thyroid cancer. Clin Cancer Res 2009;15:7119-23. [PubMed]
- Tran TN, Selinger CI, Kohonen-Corish MR, et al. Fibroblast Growth Factor Receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer 2013. [Epub ahead of print]. [PubMed]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116-29. [PubMed]
- Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One 2011;6:e20351. [PubMed]
- Ikeda K, Wang L, Torres R, et al. Discoidin domain receptor 2 interacts with Src and Shc following its activation by type I collagen. J Biol Chem 2002;277:19206-12. [PubMed]
- Knudson AG. Antioncogenes and human cancer. Proc Natl Acad Sci USA 1993;90:10914-21. [PubMed]
- Raso MG, Wistuba II. Molecular pathogenesis of early-stage non-small cell lung cancer and a proposal for tissue banking to facilitate identification of new biomarkers. J Thorac Oncol 2007;2:S128-35. [PubMed]
- Mogi A, Kuwano H. TP53 mutations in nonsmall cell lung cancer. J Biomed Biotech 2011;2011:583929.
- Wistuba II, Berry J, Behrens C, et al. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res 2000;6:2604-10. [PubMed]
- D’Amico D, Carbone D, Mitsudomi T, et al. High frequency of somatically aquired p53 mutatinos in small-cell lung cancer cell lines and tumors. Oncogene 1992;7:339-46. [PubMed]
- Tammemagi MC, McLaughlin JR, Bull SB. Meta-analyses of p53 tumor suppressor gene alterations and clinicopathological features in resected lung cancers. Cancer Epidemiol Biomarkers Prev 1999;8:625-34. [PubMed]
- Husgafvel-Pursiainen K, Boffetta P, Kannio A, et al. p53 mutations and exposure to environmental tobacco mmoke in a multicenter study on lung Cancer. Cancer Res 2000;60:2906-11. [PubMed]
- Takagi Y, Osada H, Kuroishi T, et al. p53 mutations in non-small-cell lung cancers occurring in individuals without a past history of actve smoking. Br J Cancer 1998;77:1568-72. [PubMed]
- Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J 2001;18:705-19. [PubMed]
- Jin G, Kim MJ, Jeon HS, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung cancer 2010;69:279-83. [PubMed]
- Marsit CJ, Zheng S, Aldape K, et al. PTEN expression in non–small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol 2005;36:768-76. [PubMed]
- Marignani PA. LKB1, the multitasking tumour suppressor kinase. J Clin Pathol 2005;58:15-9. [PubMed]
- Shaw RJ, Bardeesy N, Manning BD, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 2004;6:91-9. [PubMed]
- Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasion but not Asian lung cancer patients. Br J Cancer 2008;99:245-52. [PubMed]
- Matsumoto S, Iwakawa R, Takahashi K, et al. Prevalence and specifcity of LKB1 genetic alterations in lung cancers. Oncogene 2007;26:5911-8. [PubMed]
- Sanchez-Cespedes M, Parrella P, Esteller M, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res 2002;62:3659-62. [PubMed]
- Onozato R, Kosaka T, Achiwa H, et al. LKB1 gene mutations in Japanese lung cancer patients. Cancer Sci 2007;98:1747-51. [PubMed]
- Harbour JW, Lai SL, Whang-Peng J, et al. Abnormalities in structure and epxression of the human retinoblastoma gene in SCLC. Science 1988;241:353-7. [PubMed]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 2002;3:11-20. [PubMed]
- Brambilla E, Moro D, Gazzeri S, et al. Alterations of the expression of Rb, p16(INK4A) and cyclin D1 in non-small cell lung carcinoma and their clinical significance. J Pathol 1999;188:351-60. [PubMed]
- Otterson GA, Kratzke RA, Coxon A, et al. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB. Oncogene 1994;9:3375-8. [PubMed]
- Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med 2012;18:349-51. [PubMed]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [PubMed]
- Sriram KB, Larsen JE, Yang IA, et al. Genomic medicine in non-small cell lung cancer: paving the path to personalized care. Respirology 2011;16:257-63. [PubMed]
- Paik PK, Johnson ML, D’Angelo SP, et al. Driver mutations determine survival in smokers and never-smokers with stage IIIB/IV lung adenocarcinomas. Cancer 2012;118:5840-7. [PubMed]
- Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res 2011;17:2081-6. [PubMed]
- Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: relevance for clinical practice and clinical trials. J Clin Oncol 2013;31:992-1001. [PubMed]
- Motoi N, Szoke J, Riely GJ, et al. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol 2008;32:810-27. [PubMed]
- Zakowski MF, Hussain S, Pao W, et al. Morphologic features of adenocarcinoma of the lung predictive of response to the epidermal growth factor receptor kinase inhibitors erlotinib and gefitinib. Arch Pathol Lab Med 2009;133:470-7. [PubMed]
- Ohtsuka K, Ohnishi H, Fujiwara M, et al. Abnormalities of epidermal growth factor receptor in lung squamous-cell carcinomas, adenosquamous carcinomas, and large-cell carcinomas: tyrosine kinase domain mutations are not rare in tumors with an adenocarcinoma component. Cancer 2007;109:741-50. [PubMed]
- Mok T, Wu YL, Zhang L. A small step towards personalized medicine for non-small cell lung cancer. Discov Med 2009;8:227-31. [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [PubMed]
- Yang JC, Shih JY, Su WC, et al. Afatinib for patients with lung adenocarcinoma and epidermal growth factor receptor mutations (LUX-Lung 2): a phase 2 trial. Lancet Oncol 2012;13:539-48. [PubMed]
- Wu JY, Yu CJ, Chang YC, et al. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res 2011;17:3812-21. [PubMed]
- Lee W, Jiang Z, Liu J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 2010;465:473-7. [PubMed]
- Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 2010;463:184-90. [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [PubMed]


